Team:Buenos Aires/Project/Schemes
From 2012.igem.org
(Difference between revisions)
(→Scheme 3: Cross-population Control) |
(→Schemes) |
||
| (29 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Buenos_Aires/Templates/menu}} | {{:Team:Buenos_Aires/Templates/menu}} | ||
| - | + | = Schemes = | |
| - | + | During the design phase of the project we though of several alternative schemes, each of which have pros and cons. Here we do a short summary of these schemes. | |
| - | In the cross-feeding scheme we plan to use strains which are auxotrophic for certain amino acids (e.g. lysine and tryptophan). By starting with a strain with both auxotrophies, A will be created by rescuing the biosynthetic pathway for Trp and forcing its export from the cell. Analogously, we will create strain B by rescuing the Lys pathway and forcing the secretion of Lys from the cell. | + | == Crossfeeding == |
| + | |||
| + | {| | ||
| + | |- | ||
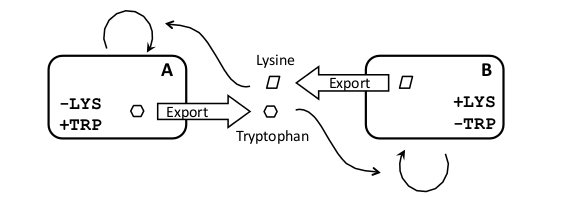
| + | |In the cross-feeding scheme we plan to use strains which are auxotrophic for certain amino acids (e.g. lysine and tryptophan). By starting with a strain with both auxotrophies, '''A''' will be created by rescuing the biosynthetic pathway for ''Trp'' and forcing its export from the cell. Analogously, we will create strain '''B''' by rescuing the ''Lys'' pathway and forcing the secretion of ''Lys'' from the cell. | ||
Amino-acid export from the cell can be engineered in the form of peptides (enriched for the relevant amino acid) which, in turn, can be easily controlled at the transcriptional and translational level. Export will be done either with secretion tags or as «trojan peptides» (Derossi et. al., 1998), which can diffuse through the plasma membrane. The main tuneability of the system could be changing the export rates for each amino acid. | Amino-acid export from the cell can be engineered in the form of peptides (enriched for the relevant amino acid) which, in turn, can be easily controlled at the transcriptional and translational level. Export will be done either with secretion tags or as «trojan peptides» (Derossi et. al., 1998), which can diffuse through the plasma membrane. The main tuneability of the system could be changing the export rates for each amino acid. | ||
Because of the availability of auxotrophic mutants in yeast, this chassis is ideally suited for the crossfeeding circuit design. | Because of the availability of auxotrophic mutants in yeast, this chassis is ideally suited for the crossfeeding circuit design. | ||
| + | |[[File:Bsas2012-schemes-crossfeeding.png|500px]] | ||
| + | |} | ||
| + | {| class="wikitable" style="width: 95%" | ||
| + | ! scope="row" style="background: #7ac5e8; width: 50%" | Pros | ||
| + | ! scope="row" style="background: #7ac5e8; width: 50%" | Cons | ||
| + | |- | ||
| + | | | ||
| + | * Relatively easy to implement, but new parts would need to be created. | ||
| + | * Feasible based on previous studies (Shou et. al., 2006). | ||
| + | * Easily scalable, as many strains as initial amino acid auxotrophies can be added. | ||
| + | | | ||
| + | * Doesn’t work at very low or very high cell densities. | ||
| + | * Only works in synthetic defined mediums. | ||
| + | * A considerable initial cell density is required to start the system. | ||
| + | |} | ||
| + | == Independent Population Control == | ||
| - | + | {| | |
| + | |- valign="top" | ||
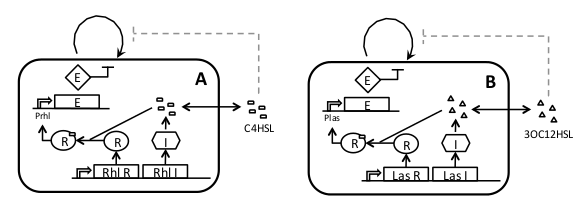
| + | |In this scheme, quorum sensing (QS) controls the density of each strain in an independent manner. | ||
| - | + | The system has already been implemented for a single strain, using the ''LuxR/LuxI'' system from ''Vibrio fischeri'', and the CcdB killer protein (E in the figure) under control of a LuxR responsive promoter (You et. Al. 2004). This strain was shown to stabilize its growth at a cell density below saturation. | |
| - | + | Our idea is to extend the system, creating new strains with the same overall functioning but different autoinducers so as to avoid interference with each other. Tuneability can be achieved by modulating both production and sensitivity to the autoinducer. | |
| + | |[[File:Bsas2012-schemes-indep-population.png|500px]] | ||
| + | |} | ||
| - | + | {| class="wikitable" style="width: 95%" | |
| + | ! scope="row" style="background: #7ac5e8; width: 50%" | Pros | ||
| + | ! scope="row" style="background: #7ac5e8; width: 50%" | Cons | ||
| + | |- | ||
| + | | | ||
| + | * The system is already implemented for one strain and can be re-utilized. | ||
| + | * Easily scalable, as many strains as independent QS systems can be added. | ||
| + | * The system can be set with a small initial inoculum of each strain. | ||
| + | | | ||
| + | * Stabilization at sub-saturation OD could be achieved. | ||
| + | * A new QS mechanisms is required for each strain. | ||
| + | |} | ||
| - | - | + | == Cross-population control == |
| - | + | {| | |
| - | + | |- valign="top" | |
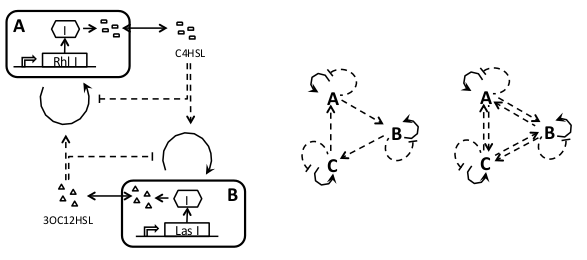
| - | - | + | |This scheme is a modification of the ''independent population control'', where the autoinducers produce some effect on the other strain(s) beside modulating the growth of the strain that produces it. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ''' | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
This coupling between strains can increase robustness and give rise to some interesting behaviors as oscillations. | This coupling between strains can increase robustness and give rise to some interesting behaviors as oscillations. | ||
| - | As more strains are added, several different connections between them can be implemented that will probably result in different properties of the system. | + | As more strains are added, several different connections between them can be implemented that will probably result in different properties of the system. |
| + | |[[File:Bsas2012-schemes-cross-population.png|500px]] | ||
| + | |} | ||
| - | + | {| class="wikitable" style="width: 95%" | |
| + | ! scope="row" style="background: #7ac5e8; width: 50%" | Pros | ||
| + | ! scope="row" style="background: #7ac5e8; width: 50%" | Cons | ||
| + | |- | ||
| + | | | ||
| + | * Required parts are available and well characterized. | ||
| + | * Stabilization at sub-saturation OD could be achieved. | ||
| + | * Should work in complex mediums. | ||
| + | * Interesting dynamics as oscillations could arise. | ||
| + | * A new QS mechanisms is required for each strain. | ||
| + | | | ||
| + | * Not scalable, modifications to several or all strains are required each time a new strain is added. | ||
| + | * A minimum OD is required for the system to work. | ||
| + | |} | ||
| - | + | == Stochastic State Transitions == | |
| - | + | {| | |
| + | |- | ||
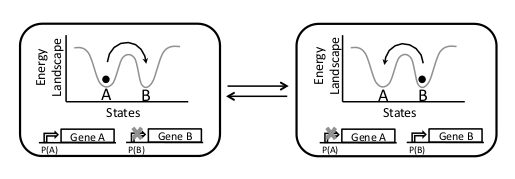
| + | |This scheme is quite different to the previous ones in the sense that there is a single strain that has several stable states (e.g. '''A''' and '''B'''). | ||
| - | + | Each state activates the transcription of different sets of genes. The transition between states is given in a stochastic manner, but the probability of transition can be externally modulated. Each cell will spend a fraction of the time in each state independently of other cell. | |
| - | + | Therefore, at any given time, cell population will be composed of a fraction of cells in each state. Tuning the transition rates between states will result in a correspondent change in the fraction of cells in each state. | |
| + | The system can be implemented as a «leaky toggle switch» | ||
| + | |[[File:Bsas2012-schemes-stochastic.png|500px]] | ||
| + | |} | ||
| - | + | {| class="wikitable" style="width: 95%" | |
| + | ! scope="row" style="background: #7ac5e8; width: 50%" | Pros | ||
| + | ! scope="row" style="background: #7ac5e8; width: 50%" | Cons | ||
| + | |- | ||
| + | | | ||
| + | * Completely independent of cell density. | ||
| + | * If a rare state induces death, a slow population growth could be achieve. | ||
| + | | | ||
| + | * All components need to be incorporated in the same strain, thus no circuit integration or isolation is possible (see applications section). | ||
| + | |} | ||
| - | + | == References == | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | # You L., Sidney Cox III R., Weiss R., Arnold1 F. H. (2004). Programmed population control by cell–cell communication and regulated killing. Nature 428, 868-871. | |
| - | + | # Derossi D., Chassaing G., Prochiantz A.. (1998). Trojan peptides: the penetratin system for intracellular delivery. Trends Cell Biol 8-2. 84-87. | |
| - | + | # Shou W., Ram S., Vilar J. M. G. (2006). Synthetic cooperation in engineered yeast populations. PNAS 104-6 1877-1882 | |
| - | + | ||
Latest revision as of 18:41, 18 September 2012

Contents |
Schemes
During the design phase of the project we though of several alternative schemes, each of which have pros and cons. Here we do a short summary of these schemes.
Crossfeeding
| Pros | Cons |
|---|---|
|
|
Independent Population Control
| Pros | Cons |
|---|---|
|
|
Cross-population control
| Pros | Cons |
|---|---|
|
|
Stochastic State Transitions
| Pros | Cons |
|---|---|
|
|
References
- You L., Sidney Cox III R., Weiss R., Arnold1 F. H. (2004). Programmed population control by cell–cell communication and regulated killing. Nature 428, 868-871.
- Derossi D., Chassaing G., Prochiantz A.. (1998). Trojan peptides: the penetratin system for intracellular delivery. Trends Cell Biol 8-2. 84-87.
- Shou W., Ram S., Vilar J. M. G. (2006). Synthetic cooperation in engineered yeast populations. PNAS 104-6 1877-1882
 "
"