Team:ETH Zurich/Modeling/Parameters
From 2012.igem.org
(Difference between revisions)
| (3 intermediate revisions not shown) | |||
| Line 23: | Line 23: | ||
|room | |room | ||
|sun*0.3, (UV<350nm)*0.05, (UV>=350nm)*0,90 | |sun*0.3, (UV<350nm)*0.05, (UV>=350nm)*0,90 | ||
| - | | | + | |assumed |
|210 W m<sup>-2</sup> | |210 W m<sup>-2</sup> | ||
|n/a | |n/a | ||
| Line 112: | Line 112: | ||
!Reference | !Reference | ||
|- | |- | ||
| - | | k_UVR8_hv||Light dependent dissociation rate UVR8 dimer|| s<sup>-1</sup>||from | + | | k_UVR8_hv||Light dependent dissociation rate UVR8 dimer|| 2.08·10<sup>-3</sup> s<sup>-1</sup>||from gels |
|- | |- | ||
| k_UVR8_decay||Dimerization rate UVR8 monomer||8.4·10<sup>-10</sup> nM<sup>-1</sup> s<sup>-1</sup>||estimate | | k_UVR8_decay||Dimerization rate UVR8 monomer||8.4·10<sup>-10</sup> nM<sup>-1</sup> s<sup>-1</sup>||estimate | ||
|- | |- | ||
| - | | KM_TetR||TetR repression coefficient ||100 nM|| | + | | KM_TetR||TetR repression coefficient ||100 nM||assumed |
|- | |- | ||
| n_TetR||TetR cooperativity coefficient||1||<span class='eth_reference'>[GarciaOjalvo2004]</span> | | n_TetR||TetR cooperativity coefficient||1||<span class='eth_reference'>[GarciaOjalvo2004]</span> | ||
|- | |- | ||
| - | | k_Ptet||Tet promoter expression strength|| | + | | k_Ptet||Tet promoter expression strength||1.1 nM s<sup>-1</sup>||optimised |
|- | |- | ||
| - | | A||Basal expression fraction||0. | + | | A||Basal expression fraction||0.05||assumed |
|- | |- | ||
| - | | n||Hill-like pABA cooperativity coefficient|| | + | | n||Hill-like pABA cooperativity coefficient||10<sup>-5</sup>||assumed |
|- | |- | ||
| - | | k_deg||Protein degradation rate|| | + | | k_deg||Protein degradation rate||3.85·10<sup>-5</sup> s<sup>-1</sup>||assumed |
|- | |- | ||
| - | | KM_PabAB||PabAB Michaelis constant|| | + | | KM_PabAB||PabAB Michaelis constant||960·10<sup>3</sup> nM||<span class='eth_reference'>[Roux1992]</span> |
|- | |- | ||
| k_cat||PabAB catalysis rate||0.65 s<sup>-1</sup>||<span class='eth_reference'>[Roux1992]</span> | | k_cat||PabAB catalysis rate||0.65 s<sup>-1</sup>||<span class='eth_reference'>[Roux1992]</span> | ||
|- | |- | ||
| - | | Chor0||Intracellular chorismate concentration|| | + | | Chor0||Intracellular chorismate concentration||100 mM||assumed |
|- | |- | ||
| - | | k_out||pABA outflux rate|| | + | | k_out||pABA outflux rate||3.85·10<sup>-4</sup> s<sup>-1</sup>||assumed |
|} | |} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
=== LovTAP-Cph8 === | === LovTAP-Cph8 === | ||
| Line 200: | Line 156: | ||
| KM_cI||cI repression coefficient||8 nM||<span class='eth_reference'>[Basu2005]</span> | | KM_cI||cI repression coefficient||8 nM||<span class='eth_reference'>[Basu2005]</span> | ||
|- | |- | ||
| - | | KM_TetR||TetR repression coefficient||100 nM|| | + | | KM_TetR||TetR repression coefficient||100 nM||assumed |
|- | |- | ||
| n_LacI||LacI cooperativity coefficient||2||<span class='eth_reference'>[Basu2005]</span> | | n_LacI||LacI cooperativity coefficient||2||<span class='eth_reference'>[Basu2005]</span> | ||
| Line 208: | Line 164: | ||
| n_TetR||TetR cooperativity coefficient||1||<span class='eth_reference'>[GarciaOjalvo2004]</span> | | n_TetR||TetR cooperativity coefficient||1||<span class='eth_reference'>[GarciaOjalvo2004]</span> | ||
|- | |- | ||
| - | | n_Cph8||Cph8 cooperativity coefficient||1|| | + | | n_Cph8||Cph8 cooperativity coefficient||1||assumed |
|- | |- | ||
| - | | n_LOV||LOV cooperativity coefficient||1|| | + | | n_LOV||LOV cooperativity coefficient||1||assumed |
|- | |- | ||
| k_LOV_decay||Dark decay rate of active LOV||5.8·10<sup>-3</sup> s<sup>-1</sup>||<span class='eth_reference'>[Drepper2007]</span> | | k_LOV_decay||Dark decay rate of active LOV||5.8·10<sup>-3</sup> s<sup>-1</sup>||<span class='eth_reference'>[Drepper2007]</span> | ||
| Line 224: | Line 180: | ||
| k_P_L||Lambda P_L expression strength||3.0·10<sup>-2</sup> nM s<sup>-1</sup>||optimized | | k_P_L||Lambda P_L expression strength||3.0·10<sup>-2</sup> nM s<sup>-1</sup>||optimized | ||
|- | |- | ||
| - | | A||Basal expression fraction||0. | + | | A||Basal expression fraction||0.05||assumed |
|- | |- | ||
| - | | k_deg||Protein degradation rate||1.9·10<sup>-3</sup> s<sup>-1</sup>|| | + | | k_deg||Protein degradation rate||1.9·10<sup>-3</sup> s<sup>-1</sup>||assumed |
|} | |} | ||
Parameters denoted with | Parameters denoted with | ||
| - | * '' | + | * ''assumed'' have been assumed from intuition |
* ''estimate'' have been calculated from gels, approximate experimental numbers or other related biological numbers | * ''estimate'' have been calculated from gels, approximate experimental numbers or other related biological numbers | ||
* ''optimized'' have been adjusted such that the constructs work optimally. These are target constraints that biologists should care for when selecting promoters. | * ''optimized'' have been adjusted such that the constructs work optimally. These are target constraints that biologists should care for when selecting promoters. | ||
| + | |||
| + | === Sun Protection Factor === | ||
| + | |||
| + | {| {{table}} | ||
| + | ! Parameter||Description||Value||Reference | ||
| + | |- | ||
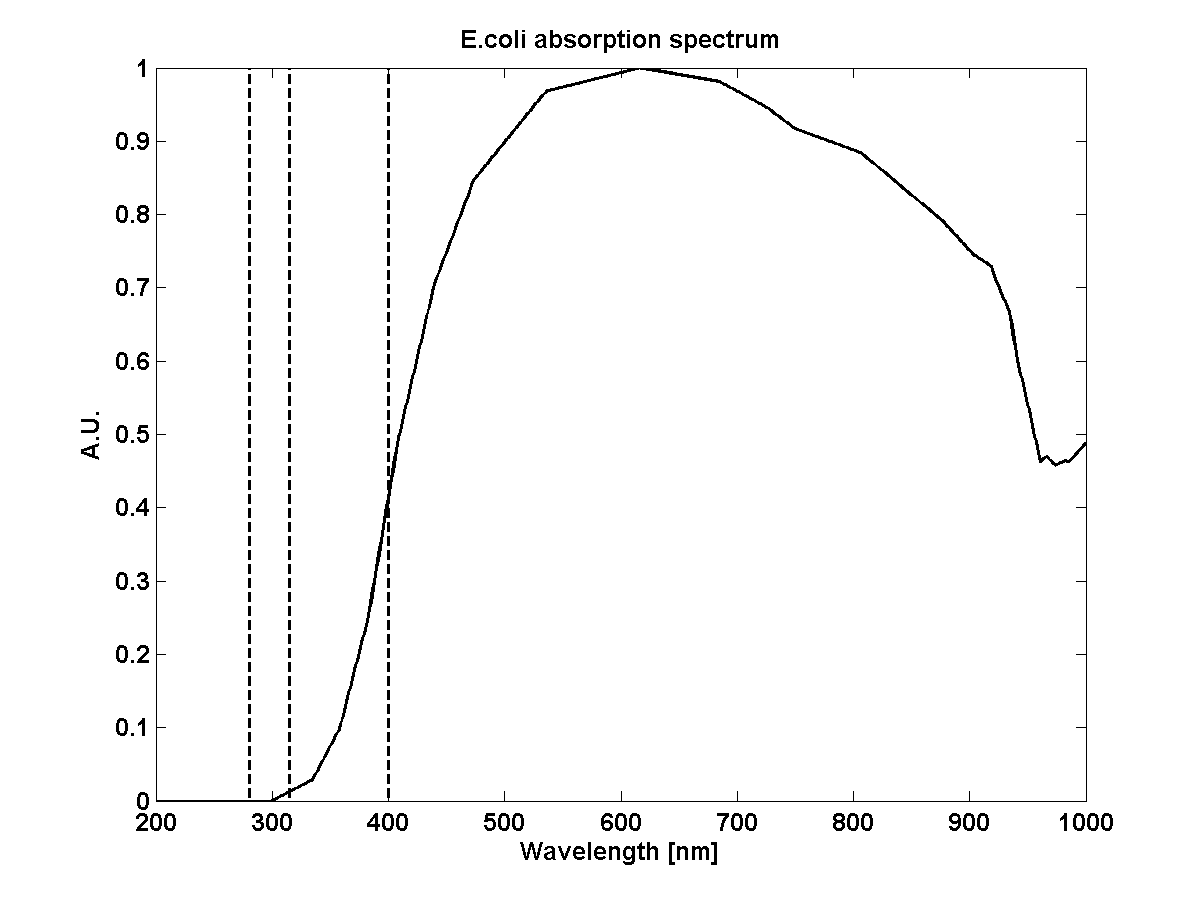
| + | | ecoli_v||Volume of E.coli||2.0e-18 m3||[http://bionumbers.hms.harvard.edu/Includes/KeyNumbersLinks.pdf Bionumbers] | ||
| + | |- | ||
| + | | ecoli_extcoeff||Extinction coefficient of E.coli at 600nm||6.022e10 m2 mol-1||Computed via OD600 | ||
| + | |- | ||
| + | | ecoli_absorption||Absorption spectrum E.coli||[[File:ETH_modeling_abs_ecoli.png|frameless|150px]]||<span class='eth_reference'>[Kiefer2010]</span> | ||
| + | |- | ||
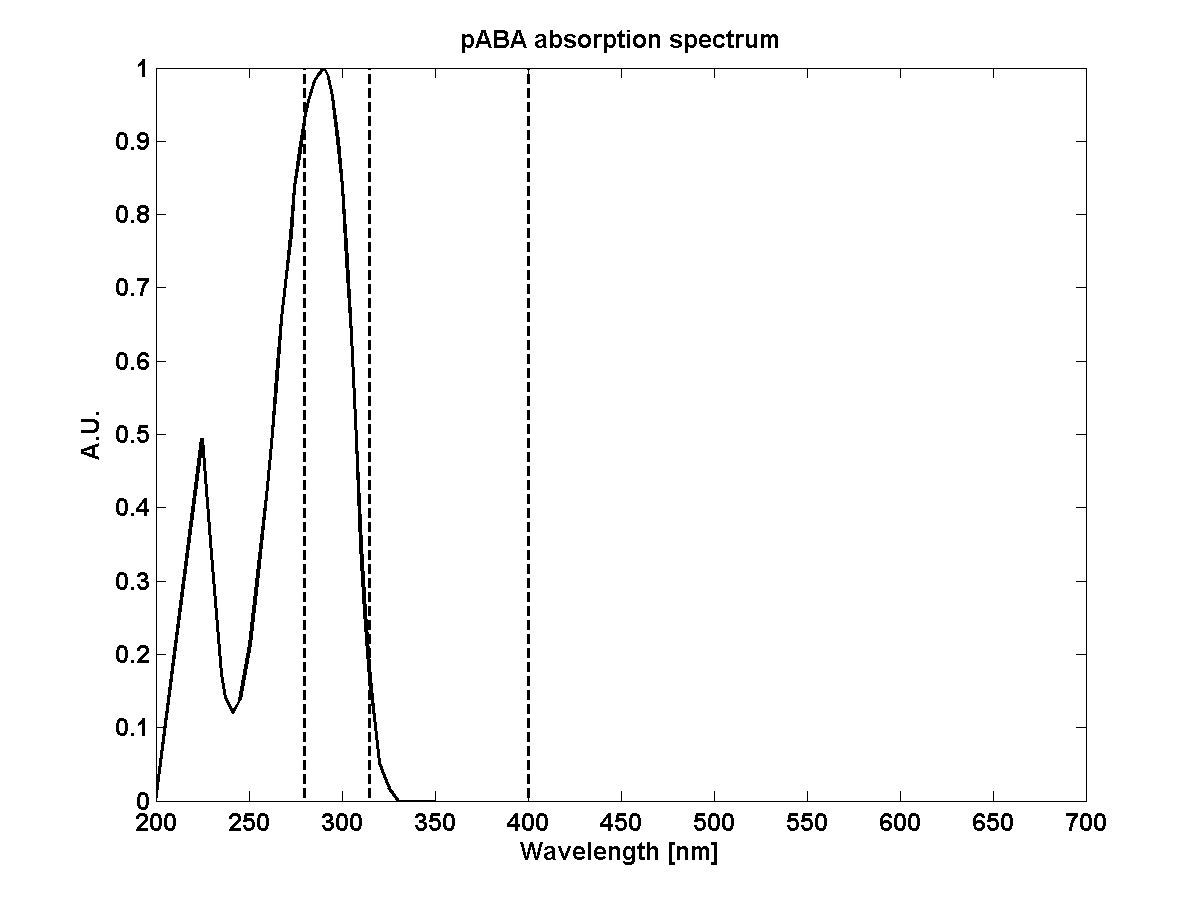
| + | | pABA_extcoeff||Extinction coefficient of pABA at 290nm||1.9e3 m2 mol-1||<span class='eth_reference'>[Quinlivan2003]</span> | ||
| + | |- | ||
| + | | pABA_absorption||Absorption spectrum pABA||[[File:ETH_modeling_abs_paba.png|frameless|150px]]||<span class='eth_reference'>[EC2006]</span> | ||
| + | |- | ||
| + | | pABA_molarweight||Molar weight of pABA||137.14 g mol-1||[http://www.sigmaaldrich.com/catalog/product/sigma/a9878?lang=de®ion=DE Datasheet] | ||
| + | |- | ||
| + | | layer_height||Height of sunscreen layer||2e-5 m||<span class='eth_reference'>[Vainio2001]</span> | ||
| + | |- | ||
| + | |} | ||
| + | |||
{{:Team:ETH_Zurich/Templates/Footer}} | {{:Team:ETH_Zurich/Templates/Footer}} | ||
Latest revision as of 23:23, 26 October 2012
Contents |
Parameters for modeling
Photoinduction
Light sources
| Name | Description | Reference | approx. flux at probe | Distance source - probe |
|---|---|---|---|---|
| sun | natural sun light | ISO 9845-1, ASTMG173 | 640 W m-2 | n/a |
| room | sun*0.3, (UV<350nm)*0.05, (UV>=350nm)*0,90 | assumed | 210 W m-2 | n/a |
| bulb200W | Incandescent light bulb | GE200Clear | 12 W m-2 | 1 m |
Photoconversion cross section
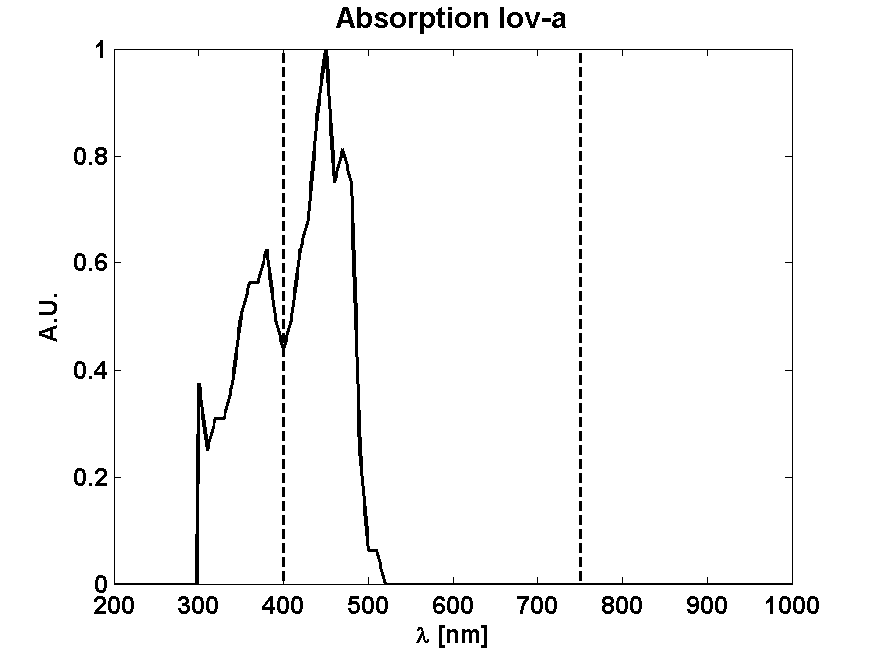
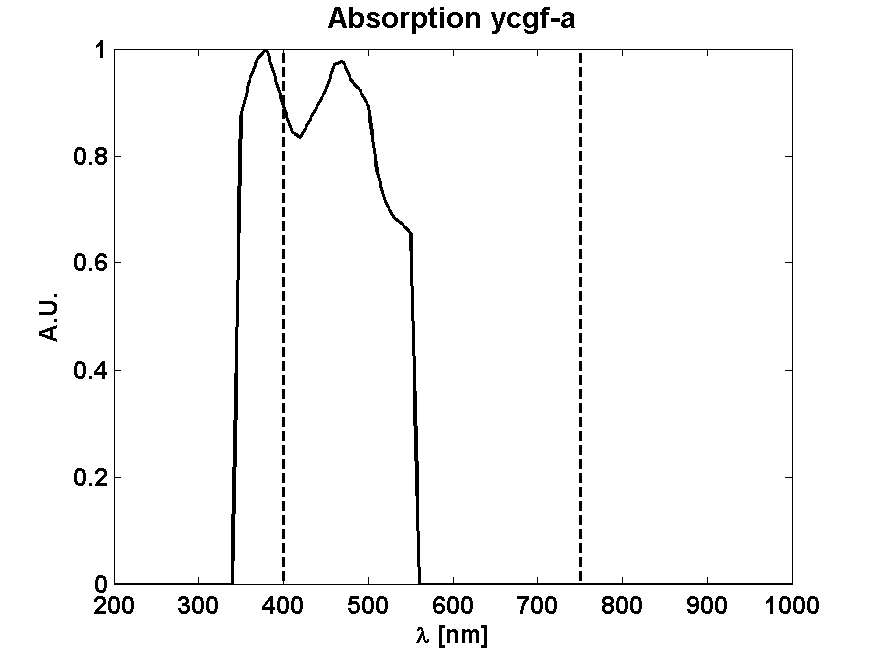
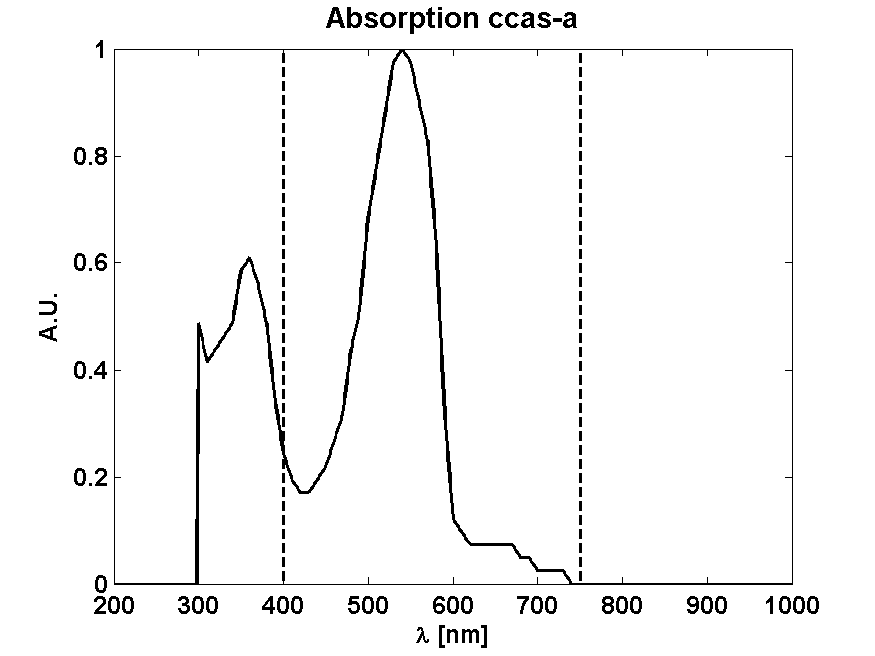
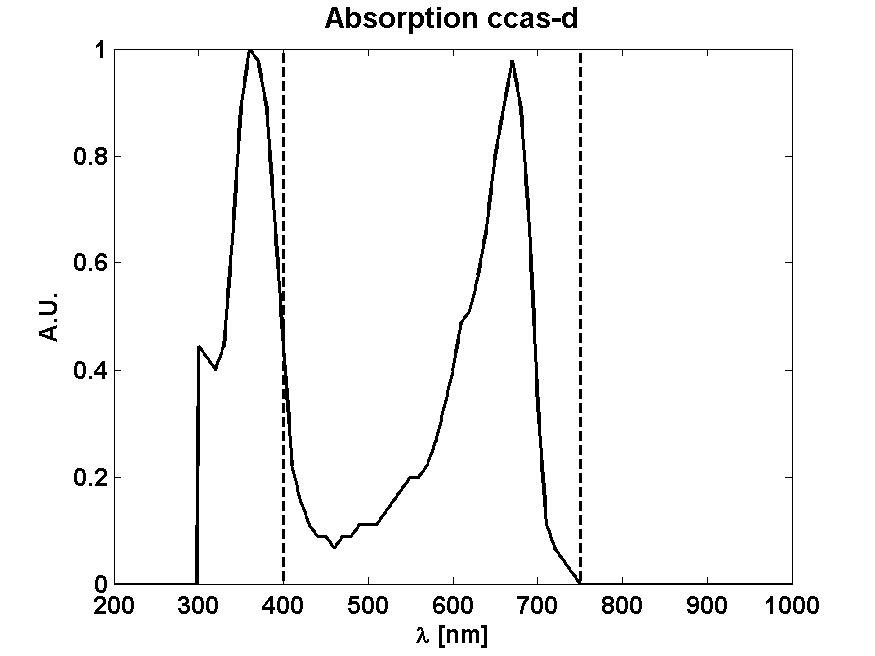
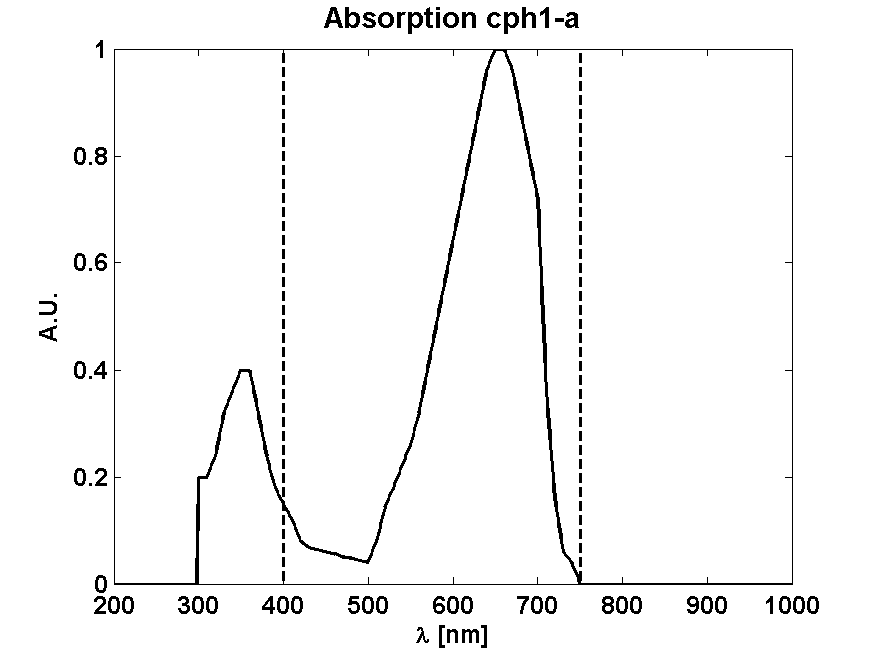
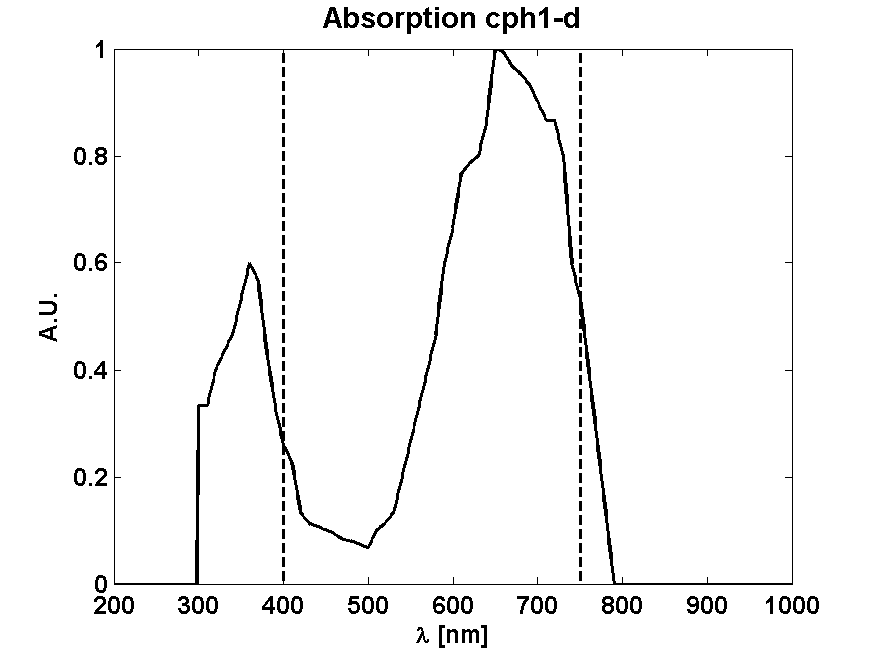
From Absorption spectrum
From Photon effectiveness
| Receptor | Activation photon effectiveness | Deactivation photon effectiveness | References |
|---|---|---|---|
| UVR8 | 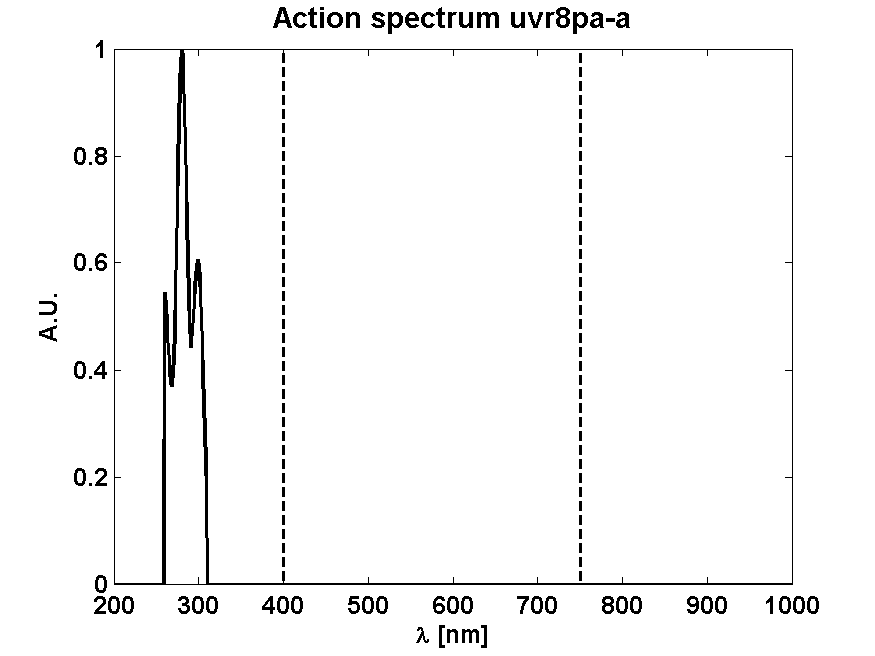
| n/a | [Brown2009] |
UVR8
| Parameter | Description | Value | Reference |
|---|---|---|---|
| k_UVR8_hv | Light dependent dissociation rate UVR8 dimer | 2.08·10-3 s-1 | from gels |
| k_UVR8_decay | Dimerization rate UVR8 monomer | 8.4·10-10 nM-1 s-1 | estimate |
| KM_TetR | TetR repression coefficient | 100 nM | assumed |
| n_TetR | TetR cooperativity coefficient | 1 | [GarciaOjalvo2004] |
| k_Ptet | Tet promoter expression strength | 1.1 nM s-1 | optimised |
| A | Basal expression fraction | 0.05 | assumed |
| n | Hill-like pABA cooperativity coefficient | 10-5 | assumed |
| k_deg | Protein degradation rate | 3.85·10-5 s-1 | assumed |
| KM_PabAB | PabAB Michaelis constant | 960·103 nM | [Roux1992] |
| k_cat | PabAB catalysis rate | 0.65 s-1 | [Roux1992] |
| Chor0 | Intracellular chorismate concentration | 100 mM | assumed |
| k_out | pABA outflux rate | 3.85·10-4 s-1 | assumed |
LovTAP-Cph8
| Parameter | Description | Value | Reference |
|---|---|---|---|
| k_Cph8_hv | Light dependent activation rate | s-1 | from photoinduction model |
| KM_LOV | LOV repression coefficient | 142 nM | [Strickland2007] |
| KM_Cph8 | Cph8 activation coefficient | 1000 nM | estimate |
| KM_LacI | LacI repression coefficient | 800 nM | [Basu2005] |
| KM_cI | cI repression coefficient | 8 nM | [Basu2005] |
| KM_TetR | TetR repression coefficient | 100 nM | assumed |
| n_LacI | LacI cooperativity coefficient | 2 | [Basu2005] |
| n_cI | cI cooperativity coefficient | 2 | [Basu2005] |
| n_TetR | TetR cooperativity coefficient | 1 | [GarciaOjalvo2004] |
| n_Cph8 | Cph8 cooperativity coefficient | 1 | assumed |
| n_LOV | LOV cooperativity coefficient | 1 | assumed |
| k_LOV_decay | Dark decay rate of active LOV | 5.8·10-3 s-1 | [Drepper2007] |
| k_Cph8_decay | Dark decay rate of active Cph8 | 5.8·10-3 s-1 | estimate |
| k_Ptrp | Trp promoter expression strength | 2.23 nM s-1 | optimized |
| k_PompC | OmpC promoter expression strength | 3.454·10-1 nM s-1 | optimized |
| k_P_R | Lambda P_R expression strength | 4.21·10-2 nM s-1 | optimized |
| k_P_L | Lambda P_L expression strength | 3.0·10-2 nM s-1 | optimized |
| A | Basal expression fraction | 0.05 | assumed |
| k_deg | Protein degradation rate | 1.9·10-3 s-1 | assumed |
Parameters denoted with
- assumed have been assumed from intuition
- estimate have been calculated from gels, approximate experimental numbers or other related biological numbers
- optimized have been adjusted such that the constructs work optimally. These are target constraints that biologists should care for when selecting promoters.
Sun Protection Factor
References
- Brown, B. a, Headland, L. R., & Jenkins, G. I. (2009). UV-B action spectrum for UVR8-mediated HY5 transcript accumulation in Arabidopsis. Photochemistry and photobiology, 85(5), 1147–55.
- Christie, J. M., Salomon, M., Nozue, K., Wada, M., & Briggs, W. R. (1999): LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proceedings of the National Academy of Sciences of the United States of America, 96(15), 8779–83.
- Christie, J. M., Arvai, A. S., Baxter, K. J., Heilmann, M., Pratt, A. J., O’Hara, A., Kelly, S. M., et al. (2012). Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science (New York, N.Y.), 335(6075), 1492–6.
- Cloix, C., & Jenkins, G. I. (2008). Interaction of the Arabidopsis UV-B-specific signaling component UVR8 with chromatin. Molecular plant, 1(1), 118–28.
- Cox, R. S., Surette, M. G., & Elowitz, M. B. (2007). Programming gene expression with combinatorial promoters. Molecular systems biology, 3(145), 145. doi:10.1038/msb4100187
- Drepper, T., Eggert, T., Circolone, F., Heck, A., Krauss, U., Guterl, J.-K., Wendorff, M., et al. (2007). Reporter proteins for in vivo fluorescence without oxygen. Nature biotechnology, 25(4), 443–5
- Drepper, T., Krauss, U., & Berstenhorst, S. M. zu. (2011). Lights on and action! Controlling microbial gene expression by light. Applied microbiology, 23–40.
- EuropeanCommission (2006). SCIENTIFIC COMMITTEE ON CONSUMER PRODUCTS SCCP Opinion on Biological effects of ultraviolet radiation relevant to health with particular reference to sunbeds for cosmetic purposes.
- Elvidge, C. D., Keith, D. M., Tuttle, B. T., & Baugh, K. E. (2010). Spectral identification of lighting type and character. Sensors (Basel, Switzerland), 10(4), 3961–88.
- GarciaOjalvo, J., Elowitz, M. B., & Strogatz, S. H. (2004). Modeling a synthetic multicellular clock: repressilators coupled by quorum sensing. Proceedings of the National Academy of Sciences of the United States of America, 101(30), 10955–60.
- Gao Q, Garcia-Pichel F. (2011). Microbial ultraviolet sunscreens. Nat Rev Microbiol. 9(11):791-802.
- Goosen N, Moolenaar GF. (2008) Repair of UV damage in bacteria. DNA Repair (Amst).7(3):353-79.
- Heijde, M., & Ulm, R. (2012). UV-B photoreceptor-mediated signalling in plants. Trends in plant science, 17(4), 230–7.
- Hirose, Y., Narikawa, R., Katayama, M., & Ikeuchi, M. (2010). Cyanobacteriochrome CcaS regulates phycoerythrin accumulation in Nostoc punctiforme, a group II chromatic adapter. Proceedings of the National Academy of Sciences of the United States of America, 107(19), 8854–9.
- Hirose, Y., Shimada, T., Narikawa, R., Katayama, M., & Ikeuchi, M. (2008). Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proceedings of the National Academy of Sciences of the United States of America, 105(28), 9528–33.
- Kast, Asif-Ullah & Hilvert (1996) Tetrahedron Lett. 37, 2691 - 2694., Kast, Asif-Ullah, Jiang & Hilvert (1996) Proc. Natl. Acad. Sci. USA 93, 5043 - 5048
- Kiefer, J., Ebel, N., Schlücker, E., & Leipertz, A. (2010). Characterization of Escherichia coli suspensions using UV/Vis/NIR absorption spectroscopy. Analytical Methods, 9660. doi:10.1039/b9ay00185a
- Kinkhabwala, A., & Guet, C. C. (2008). Uncovering cis regulatory codes using synthetic promoter shuffling. PloS one, 3(4), e2030.
- Krebs in Deutschland 2005/2006. Häufigkeiten und Trends. 7. Auflage, 2010, Robert Koch-Institut (Hrsg) und die Gesellschaft der epidemiologischen Krebsregister in Deutschland e. V. (Hrsg). Berlin.
- Lamparter, T., Michael, N., Mittmann, F., & Esteban, B. (2002). Phytochrome from Agrobacterium tumefaciens has unusual spectral properties and reveals an N-terminal chromophore attachment site. Proceedings of the National Academy of Sciences of the United States of America, 99(18), 11628–33.
- Levskaya, A. et al (2005). Engineering Escherichia coli to see light. Nature, 438(7067), 442.
- Mancinelli, A. (1986). Comparison of spectral properties of phytochromes from different preparations. Plant physiology, 82(4), 956–61.
- Nakasone, Y., Ono, T., Ishii, A., Masuda, S., & Terazima, M. (2007). Transient dimerization and conformational change of a BLUF protein: YcgF. Journal of the American Chemical Society, 129(22), 7028–35.
- Orth, P., & Schnappinger, D. (2000). Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nature structural biology, 215–219.
- Parkin, D.M., et al., Global cancer statistics, 2002. CA: a cancer journal for clinicians, 2005. 55(2): p. 74-108.
- Rajagopal, S., Key, J. M., Purcell, E. B., Boerema, D. J., & Moffat, K. (2004). Purification and initial characterization of a putative blue light-regulated phosphodiesterase from Escherichia coli. Photochemistry and photobiology, 80(3), 542–7.
- Rizzini, L., Favory, J.-J., Cloix, C., Faggionato, D., O’Hara, A., Kaiserli, E., Baumeister, R., et al. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science (New York, N.Y.), 332(6025), 103–6.
- Roux, B., & Walsh, C. T. (1992). p-aminobenzoate synthesis in Escherichia coli: kinetic and mechanistic characterization of the amidotransferase PabA. Biochemistry, 31(30), 6904–10.
- Strickland, D. (2008). Light-activated DNA binding in a designed allosteric protein. Proceedings of the National Academy of Sciences of the United States of America, 105(31), 10709–10714.
- Sinha RP, Häder DP. UV-induced DNA damage and repair: a review. Photochem Photobiol Sci. (2002). 1(4):225-36
- Sambandan DR, Ratner D. (2011). Sunscreens: an overview and update. J Am Acad Dermatol. 2011 Apr;64(4):748-58.
- Tabor, J. J., Levskaya, A., & Voigt, C. A. (2011). Multichromatic Control of Gene Expression in Escherichia coli. Journal of Molecular Biology, 405(2), 315–324.
- Thibodeaux, G., & Cowmeadow, R. (2009). A tetracycline repressor-based mammalian two-hybrid system to detect protein–protein interactions in vivo. Analytical biochemistry, 386(1), 129–131.
- Tschowri, N., & Busse, S. (2009). The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes & development, 522–534.
- Tschowri, N., Lindenberg, S., & Hengge, R. (2012). Molecular function and potential evolution of the biofilm-modulating blue light-signalling pathway of Escherichia coli. Molecular microbiology.
- Tyagi, A. (2009). Photodynamics of a flavin based blue-light regulated phosphodiesterase protein and its photoreceptor BLUF domain.
- Vainio, H. & Bianchini, F. (2001). IARC Handbooks of Cancer Prevention: Volume 5: Sunscreens. Oxford University Press, USA
- Quinlivan, Eoin P & Roje, Sanja & Basset, Gilles & Shachar-Hill, Yair & Gregory, Jesse F & Hanson, Andrew D. (2003). The folate precursor p-aminobenzoate is reversibly converted to its glucose ester in the plant cytosol. The Journal of biological chemistry, 278.
- van Thor, J. J., Borucki, B., Crielaard, W., Otto, H., Lamparter, T., Hughes, J., Hellingwerf, K. J., et al. (2001). Light-induced proton release and proton uptake reactions in the cyanobacterial phytochrome Cph1. Biochemistry, 40(38), 11460–71.
- Wegkamp A, van Oorschot W, de Vos WM, Smid EJ. (2007 )Characterization of the role of para-aminobenzoic acid biosynthesis in folate production by Lactococcus lactis. Appl Environ Microbiol. Apr;73(8):2673-81.
 "
"