Team:Cambridge/Lab book/Week 7
From 2012.igem.org
(→Friday (10/08/12)) |
(→Saturday (11/08/12)) |
||
| (8 intermediate revisions not shown) | |||
| Line 13: | Line 13: | ||
!align="center"|[[Team:Cambridge/Lab_book/Week_11|11]] | !align="center"|[[Team:Cambridge/Lab_book/Week_11|11]] | ||
!align="center"|[[Team:Cambridge/Lab_book/Week_12|12]] | !align="center"|[[Team:Cambridge/Lab_book/Week_12|12]] | ||
| + | !align="center"|[[Team:Cambridge/Lab_book/Week_13|13]] | ||
| + | !align="center"|[[Team:Cambridge/Lab_book/Week_14|14]] | ||
|} | |} | ||
| Line 95: | Line 97: | ||
*2nd attempt at amplifying the Lux vector in two fragments, similar to one attempted yesterday | *2nd attempt at amplifying the Lux vector in two fragments, similar to one attempted yesterday | ||
| - | |||
| - | |||
*Results: no bands of the correct size. There seems to be DNA in the well which indicates presence of very large pieces of DNA. | *Results: no bands of the correct size. There seems to be DNA in the well which indicates presence of very large pieces of DNA. | ||
| Line 128: | Line 128: | ||
---- | ---- | ||
| - | * | + | * [[Team:Cambridge/Protocols/SOB|SOB]] made up. |
'''[[Team:Cambridge/Protocols/PCRProtocol|PCR of positive control fragments]]''' | '''[[Team:Cambridge/Protocols/PCRProtocol|PCR of positive control fragments]]''' | ||
| Line 139: | Line 139: | ||
:* pSB4K5 backbone which confers kanamycin resistance (~3.3kb) | :* pSB4K5 backbone which confers kanamycin resistance (~3.3kb) | ||
| - | |||
| - | |||
*Results: successful amplification; sfGFP (lanes 1-3); pSB4K5 (lanes 4-6) | *Results: successful amplification; sfGFP (lanes 1-3); pSB4K5 (lanes 4-6) | ||
| Line 149: | Line 147: | ||
* Split mOrange vector amplification attempted again. | * Split mOrange vector amplification attempted again. | ||
| - | |||
| - | |||
*Results: no bands again. | *Results: no bands again. | ||
| Line 175: | Line 171: | ||
===Saturday (11/08/12)=== | ===Saturday (11/08/12)=== | ||
| - | + | ||
'''[[Team:Cambridge/Protocols/RestrictionDigest|Restriction digest of Yale plasmid]]''' | '''[[Team:Cambridge/Protocols/RestrictionDigest|Restriction digest of Yale plasmid]]''' | ||
| Line 183: | Line 179: | ||
*Cells from electroporation of e.coli with plasmid from Yale (08/08/12) [[Team:Cambridge/Protocols/MiniPrep|miniprepped]] to extract plasmid DNA. | *Cells from electroporation of e.coli with plasmid from Yale (08/08/12) [[Team:Cambridge/Protocols/MiniPrep|miniprepped]] to extract plasmid DNA. | ||
| - | *DNA digested with | + | *DNA digested with XhoI and SalI, expected fragment sizes 5580, 3139, 1437 |
| - | *Gel run, | + | *Gel run, results not shown as gel was very 'messy'. Gel had probably already begun to set when poured and so had many artefacts when imaged. As a result each lane required different brightness and contrast settings to visualise and so the full gel was not clearly visible on the printout. However, all of the bands could be identified on the screen and were exactly as expected and very clear and pure. |
*Bands in correct places, indicating that expected plasmid was present in the cells. This indicates that, although they work at a very low efficiency, our electro-competent cells still work. It may be valuable to use these when trying to grow up plasmids that we already have at high concentration. | *Bands in correct places, indicating that expected plasmid was present in the cells. This indicates that, although they work at a very low efficiency, our electro-competent cells still work. It may be valuable to use these when trying to grow up plasmids that we already have at high concentration. | ||
| Line 216: | Line 212: | ||
*sfGFP fragments have produced successful Gibson products in the past, so this will act as our positive control. If cells grow with the plasmid and fluoresce properly, we will know the master mix works. Otherwise, we will remake the master mix. | *sfGFP fragments have produced successful Gibson products in the past, so this will act as our positive control. If cells grow with the plasmid and fluoresce properly, we will know the master mix works. Otherwise, we will remake the master mix. | ||
| + | |||
| + | *Results: there is growth on the ampicillin plates but very little- we suspect a problem with positive control fragments DNA concentration being too low. (13/8/12) | ||
{{Template:Team:Cambridge/CAM_2012_TEMPLATE_FOOT}} | {{Template:Team:Cambridge/CAM_2012_TEMPLATE_FOOT}} | ||
Latest revision as of 03:18, 27 September 2012
| Week: | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 |
|---|
Contents |
Monday (06/08/12)
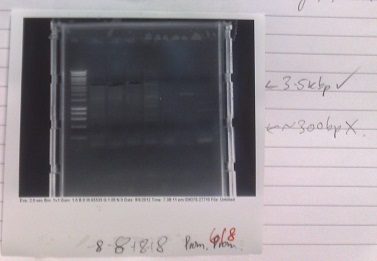
Ribosense & Existing Biobricks: PCR of Magnesium riboswitch vector fragment B and Magnesium promoter
- Normal PCR settings used, annealing temperature 57 °C, elongation step 90s long.
- Lane 5 accidentally loaded with a DNA ladder instead of loading dye.
- Expected fragment sizes:
- Lane 2-5: 3kbp
- Lane 6-7: 300bp
- After electrophoresis, found vector products had, for the most part, worked. Promoter elements were not amplified - no band of the expected size was observed.
- Positive control also failed, although this has ceased to work for several days, potentially due to DNA degradation.
Ratiometrica-Lux: Split PCR of Lux vector
- Primers for spliting pSB1C3 arrived and were used to PCR the Ratiometrica-Lux vector
- Fragment A: Vec FWD and Vec split RVS (expected size: 5kb)
- Fragment b: Vec split FWD and Vec RVS (expected size: 4.6kb)
- PCR Programme:
- Results: other than one very ambiguous band (lane 6) which might be of the right size, the others did not work.
Tuesday (07/08/12)
Production of electrocompetent e.coli
Ribosense: Gibson assembly of magnesium riboswitch
- NAD+ added to isothermal buffer*5 mix
- Gel slices from yesterday (of vector fragment B) purified.
- DNA added as follows:
- Without 8 codon substitution:
- Reaction 1: Tube 2 (07/08/12) (fragment B), Tube 1 (05/08/12) (fragment A), Tube 28 (29/08/12) (riboswitch DNA).
- Reaction 2: Tube 3 (07/08/12) (fragment B), Tube 2 (05/08/12) (fragment A), Tube 29 (29/08/12) (riboswitch DNA).
- With 8 codon substitution:
- Reaction 3: Tube 4 (07/08/12) (fragment B), Tube 1 (05/08/12) (fragment A), Tube 2 (18/07/12) (riboswitch DNA).
Ratiometrica-Flu: Gibson assembly of Fluorescent construct
- 6-piece Gibson assembly done in triplicate
- Reaction 1: Tube 1 (Fragment A), Tube 4 (Fragment B), Tube 10 (CFP), Tube 13 (B0015), Tube 16 (K143053), Tube 19 (YFP)
- Reaction 2: Tube 2 (Fragment A), Tube 5 (Fragment B), Tube 11 (CFP), Tube 14 (B0015), Tube 17 (K143053), Tube 20 (YFP)
- Reaction 3: Tube 3 (Fragment A), Tube 6 (Fragment B), Tube 12 (CFP), Tube 15 (B0015), Tube 18 (K143053), Tube 21 (YFP)
Ratiometrica-Lux: colony PCR of Lux vector
- 2nd attempt at amplifying the Lux vector in two fragments, similar to one attempted yesterday
- Results: no bands of the correct size. There seems to be DNA in the well which indicates presence of very large pieces of DNA.
Wednesday (08/08/12)
Ratiometrica & Ribosense: Electrical transformation of competent e.coli with Gibson products
- Gibson constructs from yesterday (fluorescent and magnesium riboswitch) transformed into e.coli produced yesterday.
- Cells plated out onto 50 μg/ml ampicillin plates. Put in incubator overnight.
Ratiometrica-Flu: Restriction Digest of Gibson products
- A restriction digest was performed to investigate if the plasmid was actually successfully constructed in the 6-piece Gibson reaction
- Restrictrion Enzymes: SalI (site at 5619), XhoI (site at 8040)
- Restriction Enzyme buffer: NEBuffer 3
- Results: No bands at all; probably because there was too little DNA.
Thursday (09/08/12)
Making chemically competent e.coli
- SOB made up.
PCR of positive control fragments
- Gibson positive control fragments are amplified using PCR
- sfGFP from sfGFP-ampR (~700bp)
- pSB4K5 backbone which confers kanamycin resistance (~3.3kb)
- Results: successful amplification; sfGFP (lanes 1-3); pSB4K5 (lanes 4-6)
Ratiometrica-Lux: PCR of Lux vector
- Split mOrange vector amplification attempted again.
- Results: no bands again.
Friday (10/08/12)
Making chemically competent e. coli
- Glycerol stocks of TOP10 are made competent, aliquoted into 1.5mL eppendorfs in two batches and stored in SOB at -80°C
- 50μl are transformed with pUC19 control and plated on LB agar plates to test for competence
Characterization of fluoride riboswitch sensitivity with β-galactosidase
- X-Gal made up to a concentration of 400 μg/ml with water.
- Fluoride of concentrations add concentrations mixed with bacterial culture (Yale construct containing) grown up overnight.
Saturday (11/08/12)
Restriction digest of Yale plasmid
- Cells from electroporation of e.coli with plasmid from Yale (08/08/12) miniprepped to extract plasmid DNA.
- DNA digested with XhoI and SalI, expected fragment sizes 5580, 3139, 1437
- Gel run, results not shown as gel was very 'messy'. Gel had probably already begun to set when poured and so had many artefacts when imaged. As a result each lane required different brightness and contrast settings to visualise and so the full gel was not clearly visible on the printout. However, all of the bands could be identified on the screen and were exactly as expected and very clear and pure.
- Bands in correct places, indicating that expected plasmid was present in the cells. This indicates that, although they work at a very low efficiency, our electro-competent cells still work. It may be valuable to use these when trying to grow up plasmids that we already have at high concentration.
Transformation of e.coli with Magnesium riboswitch construct
- Gibson products from 07/08/12 transformed into chemically competent cells.
- Transformants plated out on 100 μg/ml ampicillin plates
Sunday (12/08/12)
Assembly of sfGFP construct from known functional DNA fragments
- Fragments provided by Fernan assembled with Gibson.
Transformation of e.coli with fluorescent construct and sfGFP positive control
- Chemically competent e.coli cells transformed with fluorescent construct Gibson product from 07/08/12.
- E.coli cells also transformed with sfGFP Gibson product made earlier today in triplicate.
- All transformants plated out on 100 μg/ml ampicillin.
- sfGFP fragments have produced successful Gibson products in the past, so this will act as our positive control. If cells grow with the plasmid and fluoresce properly, we will know the master mix works. Otherwise, we will remake the master mix.
- Results: there is growth on the ampicillin plates but very little- we suspect a problem with positive control fragments DNA concentration being too low. (13/8/12)
 "
"