Team:UT-Tokyo/Project/Inhibition/System
From 2012.igem.org
| (3 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | {{:Team:UT-Tokyo/Template/Header|fullpagename=|subpagename=Inhibition without Knockout:<br /> System}} | + | {{:Team:UT-Tokyo/Template/Header|fullpagename=|subpagename=Inhibition without Knockout:<br /> Background & System}} |
<!-- subpagename=の後の「ページ名」部分に、このページの名称を記述してください。 --> | <!-- subpagename=の後の「ページ名」部分に、このページの名称を記述してください。 --> | ||
<html> | <html> | ||
| Line 9: | Line 9: | ||
== Background == | == Background == | ||
| - | In project | + | In project H<sub>2</sub> ''E.coli'', we tried to improve the formic acid-hydrogen pathway by overexpressing a gene ''fhlA'' which controls a step in this pathway. |
| - | However, E.coli possess a protein called HycA coded in their genome preventing unrestricted hydrogen synthesis. In order to increase the amount of hydrogen produced, it was thought that there was a need for systems that inhibit the action of negative factors such as HycA. | + | However, ''E.coli'' possess a protein called HycA coded in their genome preventing unrestricted hydrogen synthesis. In order to increase the amount of hydrogen produced, it was thought that there was a need for systems that inhibit the action of negative factors such as HycA. |
| - | In general, in order to suppress negative factors such as HycA, a gene knockout method is used. However, because it is easier to introduce a plasmid rather than knocking out a gene, we decided to try to suppress these factors by adding Biobrick Parts. A plasmid containing a large number of repeats of the binding site of the factor is introduced into E.coli as a decoy to bind the factors. Thus, the factors bind to the decoy Plasmid and we can reduce the binding to its original binding site to inhibit the function. Additionally, by changing the number of decoy binding sites, transcription can be adjusted stepwise. | + | In general, in order to suppress negative factors such as HycA, a gene knockout method is used. However, because it is easier to introduce a plasmid rather than knocking out a gene, we decided to try to suppress these factors by adding Biobrick Parts. A plasmid containing a large number of repeats of the binding site of the factor is introduced into ''E.coli'' as a decoy to bind the factors. Thus, the factors bind to the decoy Plasmid and we can reduce the binding to its original binding site to inhibit the function. Additionally, by changing the number of decoy binding sites, transcription can be adjusted stepwise. |
| - | To this end we used LacI and ArgR whose binding sites are known, and introduced multiple copies of their binding sites into plasmids to examine how the expression levels of genes downstream of these proteins were affected. | + | To this end we used LacI and ArgR whose binding sites are known, and introduced multiple copies of their binding sites into plasmids to examine how the expression levels of genes downstream of these proteins were affected. |
Latest revision as of 02:31, 27 September 2012
Inhibition without Knockout:
Background & System

Here we explain the Background & System of Inhibition without Knockout project.
Background
In project H2 E.coli, we tried to improve the formic acid-hydrogen pathway by overexpressing a gene fhlA which controls a step in this pathway.
However, E.coli possess a protein called HycA coded in their genome preventing unrestricted hydrogen synthesis. In order to increase the amount of hydrogen produced, it was thought that there was a need for systems that inhibit the action of negative factors such as HycA.
In general, in order to suppress negative factors such as HycA, a gene knockout method is used. However, because it is easier to introduce a plasmid rather than knocking out a gene, we decided to try to suppress these factors by adding Biobrick Parts. A plasmid containing a large number of repeats of the binding site of the factor is introduced into E.coli as a decoy to bind the factors. Thus, the factors bind to the decoy Plasmid and we can reduce the binding to its original binding site to inhibit the function. Additionally, by changing the number of decoy binding sites, transcription can be adjusted stepwise.
To this end we used LacI and ArgR whose binding sites are known, and introduced multiple copies of their binding sites into plasmids to examine how the expression levels of genes downstream of these proteins were affected.
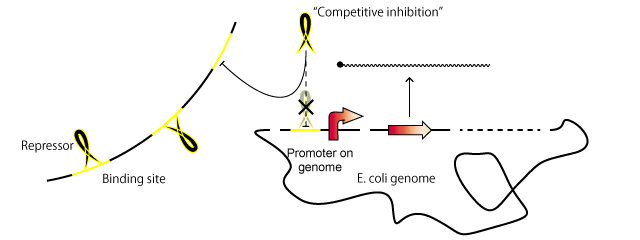
System
A repressor normally binds to a promoter and hinders transcription. By introducing extra copies of the repressor binding site in the promoter, the repressor is sequestered by these copies and the probability that the repressor binds to the promoter is decreased. For this reason, the binding of the repressor to the promoter is competitively inhibited by the repressor binding sites.
 "
"