|
|
| Line 29: |
Line 29: |
| | Starting from [http://partsregistry.org/Part:BBa_K743006| K743006] as a plasmid backbone, we were able to build our final LuxAB constructs for the bacterial luciferase using 2 different versions available at the registry (from [http://partsregistry.org/Part:BBa_K743014 Photorhabdus luminiscent, BBa_K743014] and [http://partsregistry.org/Part:BBa_K743015 Vibrio fisherii, BBa_K743015]) under an endogenous Synechocystis's promoter (transaldolase Reference???). Resulting constructs were verified by digestion (see gel images below) and corroborated by sequencing. | | Starting from [http://partsregistry.org/Part:BBa_K743006| K743006] as a plasmid backbone, we were able to build our final LuxAB constructs for the bacterial luciferase using 2 different versions available at the registry (from [http://partsregistry.org/Part:BBa_K743014 Photorhabdus luminiscent, BBa_K743014] and [http://partsregistry.org/Part:BBa_K743015 Vibrio fisherii, BBa_K743015]) under an endogenous Synechocystis's promoter (transaldolase Reference???). Resulting constructs were verified by digestion (see gel images below) and corroborated by sequencing. |
| | | | |
| - | [[File: LuxABxl_digestion.jpg| 400px| left]] | + | [[File: LuxABxl_digestion.jpg| 400px| right]] |
| - | [[File: LuxABvf_digestion.jpg| 368px| right]] | + | [[File: LuxABvf_digestion.jpg| 368px| left]] |
| | | | |
| | | | |
| | + | <br> |
| | + | <br> |
| | + | <br> |
| | + | <br> |
| | <br> | | <br> |
| | <br> | | <br> |

Plasmid Construction
According to our experimental strategy, we have built two plasmid backbones on which to insert our Lux constructs.
pSB1C3_IntK
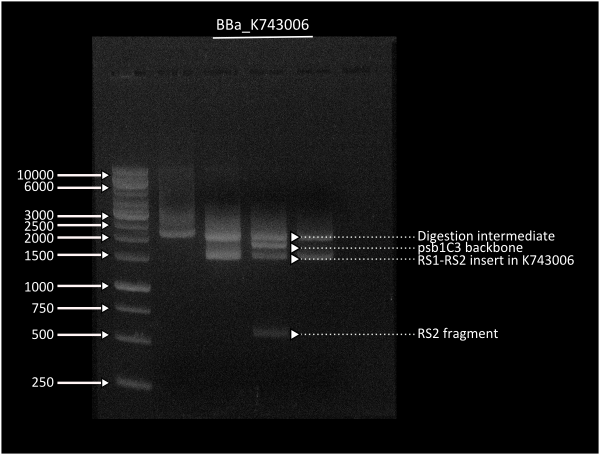
This plasmid backbone was constructed by standard assembly techniques. Recombination sites ( [http://partsregistry.org/Part:BBa_K743000 Part:BBa_K743000] and [http://partsregistry.org/Part:BBa_K743001 Part:BBa_K743001] ) were Biobricked by gibson assembly, which were consenquently ligated to neighboring parts B0015 and KanR in parallel assembly. Afterwards, we ligated both composites between them through a standard assembly reaction. Final construct was validated by digestion (see gel image at right) and corroborated through sequencing.
After obtaining [http://partsregistry.org/Part:BBa_K743006| K743006], we proceeded assembling our constructs through Gibson Assemblies.
If you wondering on why the plasmid was constructed by standard assembly and not by gibson assembly from scratch, please refer to Gibson assembly for small parts
Luciferase(s)
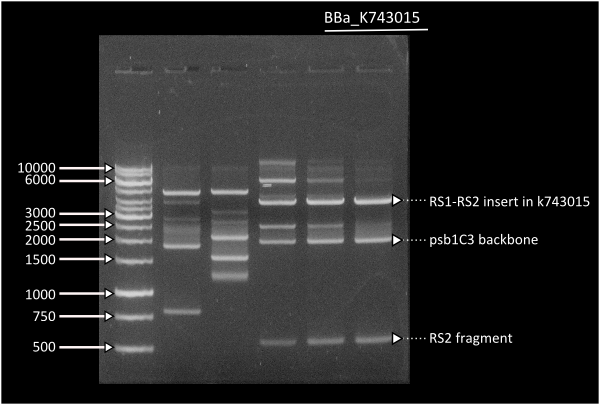
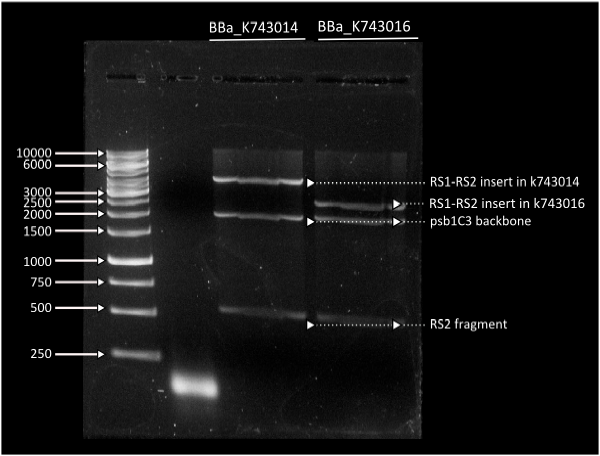
Starting from [http://partsregistry.org/Part:BBa_K743006| K743006] as a plasmid backbone, we were able to build our final LuxAB constructs for the bacterial luciferase using 2 different versions available at the registry (from [http://partsregistry.org/Part:BBa_K743014 Photorhabdus luminiscent, BBa_K743014] and [http://partsregistry.org/Part:BBa_K743015 Vibrio fisherii, BBa_K743015]) under an endogenous Synechocystis's promoter (transaldolase Reference???). Resulting constructs were verified by digestion (see gel images below) and corroborated by sequencing.
pSB1C3_IntS
After we reconsidered about using pSB1A3_IntC we designed a new plasmid backbone to place the LuxCDEG (substrate regeneration part of the Lux operon) under the Synechocystis's promoters we choose through our modelling (LINK HERE). We have obtained the plasmid backbone including all parts through Gibson Assembly and the plasmid has validated through digestion and was corroborated through sequencing.
Substrate production under Pcaa3
We have assembled all parts of the substrate production pathway ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K325902| LuxCD] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K325903| LuxEG]) for the final construct under the Pcaa3 promoter (which has also been Biobricked with code [http://partsregistry.org/wiki/index.php?title=Part:BBa_K743002 K743002] and we are now verifying for positive assembles.
Substrate production under PsigE
As this assembly was unsuccessful previously, we are re-amplifying the LuxCD part (to obtain higher concentrations of the insert) and we will retry the assembly after the wiki freeze.
Synechocystis
Transforming Synechocystis PCC 6803
(At the right side of the screen a 1 day old transformation plate of Synechocystis PCC. 6803 is shown)
After corroborating our constructs we proceded with the Synechocystis transformation for each of the constructs as soon as we obtained sufficient concentration (10ug) of the construct.
Transformation in Synechocystis takes about 2 weeks to reveal transformant colonies, which in turn have to restreaked again before PCR verification. This makes working in Synechocystis pressing as mistakes can take long to unravel.
LuxABxl under transaldolase promoter transformation
This picture shows the transformation of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K743014 K743014] after 14 days. On the left there is the control transformation plate (no DNA) on Kanamycin 25ug/mL and on the right the succesful transformation of Synechocystis colonies growing througout the Millipore membrane on the same antibiotic.
LuxABvf under transaldolase promoter transformation
It is important to note that to our knowledge we are the first iGEM team to do a succesful direct transformation of Synechocystis PCC. 6803 with naked DNA.
sfGFP with LVA tag for describing circadian behaviour
To describe the circadian behaviour of the promoter we built a fast-degrading reporter consisting of sfGPF I746916 with a LVA degradation tag in the C-terminal end of the protein. This construct will serve as a real-time reporter of promoter activity, you may find more information about the half-life of proteins with the LVA tag [http://partsregistry.org/wiki/index.php?title=Part:BBa_M0050 | here]. The construct has been verified by digestion and corroborated by sequencing.
 "
"