Team:UC Chile2/Cyanolux/Biobricks
From 2012.igem.org
(Difference between revisions)
| (9 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
<html><img src="https://static.igem.org/mediawiki/2012/d/d0/UC_Chile-C1.1.jpg" align="right" width="800"><p style="width : 200px;"> | <html><img src="https://static.igem.org/mediawiki/2012/d/d0/UC_Chile-C1.1.jpg" align="right" width="800"><p style="width : 200px;"> | ||
Integrative plasmid designed for the expression of RFP under synechocystis psbAB promoter also with a kanamycin resistance cassette for selection in cyanobacteria. The construct is flanked by neutral recombination sites RS1 and RS2, homologous to synechocystis chromosome.</p></html> | Integrative plasmid designed for the expression of RFP under synechocystis psbAB promoter also with a kanamycin resistance cassette for selection in cyanobacteria. The construct is flanked by neutral recombination sites RS1 and RS2, homologous to synechocystis chromosome.</p></html> | ||
| - | |||
| - | + | ||
| - | <p>C1.2 | + | |
| + | |||
| + | |||
| + | <html><img src="https://static.igem.org/mediawiki/2012/b/b9/UC_Chile-C1.2.jpg" align="right" width="800"><p style="width : 200px;"> | ||
| + | C1.2 | ||
This integrative plasmid is designed for the expression of RFP under synechocystis psaA2 promoter with a kanamycin resistance cassette for selection in cyanobacteria. The construct is flanked by neutral recombination sites RS1 and RS2, homologous to synechocystis chromosome. | This integrative plasmid is designed for the expression of RFP under synechocystis psaA2 promoter with a kanamycin resistance cassette for selection in cyanobacteria. The construct is flanked by neutral recombination sites RS1 and RS2, homologous to synechocystis chromosome. | ||
| - | </p> | + | </p></html> |
| - | + | ||
| - | <p>C2.1 | + | |
| - | Construct designed from pPMQAK1 expression plasmid, which replicates both in E. coli and cyanobacteria and is biobrick-compatible. It codes for RFP under synechocystis psbAB promoter.</p> | + | |
| - | + | ||
| - | <p>C2.2 | + | |
| - | Construct designed from pPMQAK1 expression plasmid, which replicates both in E. coli and cyanobacteria and is biobrick-compatible. It codes for RFP under synechocystis psaA2 promoter.</p> | + | |
| - | + | ||
| - | <p>C3 | + | |
| - | Construct designed from pPMQAK1 expression plasmid, which replicates both in E. coli and cyanobacteria and is biobrick-compatible. It codes for RFP under synechocystis psbAB promoter and cuts out the whole Ccdb toxin gene.</p> | + | |
| - | + | <html><img src="https://static.igem.org/mediawiki/2012/4/48/UC_Chile-C2.1.jpg" align="right" width="800"><p style="width : 200px;"> | |
| + | C2.1 | ||
| + | Construct designed from pPMQAK1 expression plasmid, which replicates both in E. coli and cyanobacteria and is biobrick-compatible. It codes for RFP under synechocystis psbAB promoter.</p></html> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <html><img src="https://static.igem.org/mediawiki/2012/c/c6/UC_Chile-C2.2.jpg" align="right" width="800"><p style="width : 200px;"> | ||
| + | C2.2 | ||
| + | Construct designed from pPMQAK1 expression plasmid, which replicates both in E. coli and cyanobacteria and is biobrick-compatible. It codes for RFP under synechocystis psaA2 promoter.</p></html> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <html><img src="https://2012.igem.org/File:UC_Chile-C3.1.jpg" align="right" width="800"><p style="width : 200px;"> | ||
| + | C3 | ||
| + | Construct designed from pPMQAK1 expression plasmid, which replicates both in E. coli and cyanobacteria and is biobrick-compatible. It codes for RFP under synechocystis psbAB promoter and cuts out the whole Ccdb toxin gene.</p></html> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <html><img src="https://2012.igem.org/File:UC_Chile-C3.2.jpg" align="right" width="800"><p style="width : 200px;"></p></html> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
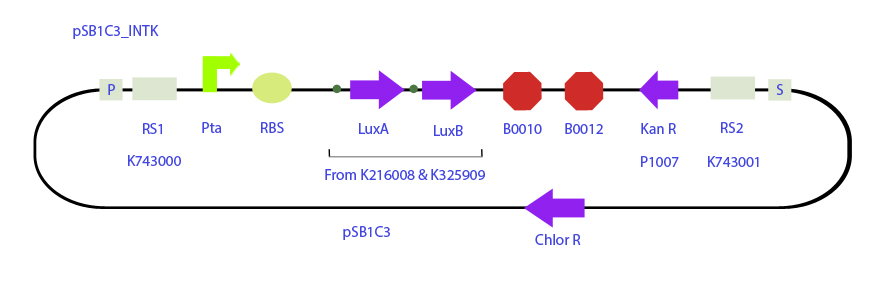
| + | <html><img src="https://2012.igem.org/File:C4.jpg" align="right" width="800"><p style="width : 200px;">Expression plasmid for the expression of LuxA and LuxB genes under psbAB promoter. | ||
| + | </p></html> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <html><img src="https://2012.igem.org/File:C5.jpg" align="right" width="800"><p style="width : 200px;">C5<br> | ||
| + | Integrative plasmid designed to integrate LuxA and LuxB and a kanamycin resistance cassette downstream the 3' end of the transaldolase gene whose mRNA levels where seen to oscillate in a circadian manner peaking at hour 14, just after dusk. It is expect the same for LuxAB mRNA levels. | ||
| + | </p></html> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Integrative plasmid designed to integrate LuxA and LuxB and a kanamycin resistance cassette downstream the 3' end of the transaldolase gene whose mRNA levels where seen to oscillate in a circadian manner peaking at hour 14, just after dusk. It is expect the same for LuxAB mRNA levels.</p> | Integrative plasmid designed to integrate LuxA and LuxB and a kanamycin resistance cassette downstream the 3' end of the transaldolase gene whose mRNA levels where seen to oscillate in a circadian manner peaking at hour 14, just after dusk. It is expect the same for LuxAB mRNA levels.</p> | ||
| Line 48: | Line 95: | ||
<br> | <br> | ||
| + | |||
| + | |||
| + | |||
| + | [[File:UC_Chile-PSB1C3_INTK.jpg|center]] | ||
| + | |||
| + | [[File:UC_Chile-PS81C3_INTS.jpg|center]] | ||
| + | |||
| + | |||
== Brief description of characterized biobricks utilized == | == Brief description of characterized biobricks utilized == | ||
Latest revision as of 00:15, 25 September 2012
 "
"