Team:METU/KillSwitch/Modelling
From 2012.igem.org
Dumankolik (Talk | contribs) |
Dumankolik (Talk | contribs) |
||
| Line 156: | Line 156: | ||
<p><b>Equations for mRNA transcription</b></p> | <p><b>Equations for mRNA transcription</b></p> | ||
| + | | ||
| | ||
| | ||
| Line 170: | Line 171: | ||
<p><img src="https://static.igem.org/mediawiki/2012/9/9f/Metu-photos-model6.png" alt="" "> | <p><img src="https://static.igem.org/mediawiki/2012/9/9f/Metu-photos-model6.png" alt="" "> | ||
</p> | </p> | ||
| + | | ||
<p><img src="https://static.igem.org/mediawiki/2012/8/8a/Metu-photos-model7.png" alt="" "> | <p><img src="https://static.igem.org/mediawiki/2012/8/8a/Metu-photos-model7.png" alt="" "> | ||
</p> | </p> | ||
| + | | ||
<p><img src="https://static.igem.org/mediawiki/2012/9/9c/Metu-photos-model8.png" alt="" "> | <p><img src="https://static.igem.org/mediawiki/2012/9/9c/Metu-photos-model8.png" alt="" "> | ||
</p> | </p> | ||
| + | | ||
<p><img src="https://static.igem.org/mediawiki/2012/4/45/Metu-photos-model9.png" alt="" "> | <p><img src="https://static.igem.org/mediawiki/2012/4/45/Metu-photos-model9.png" alt="" "> | ||
</p> | </p> | ||
| + | | ||
<p><img src="https://static.igem.org/mediawiki/2012/1/12/Metu-photos-model10.png" alt="" "> | <p><img src="https://static.igem.org/mediawiki/2012/1/12/Metu-photos-model10.png" alt="" "> | ||
</p> | </p> | ||
| + | | ||
<p><img src="https://static.igem.org/mediawiki/2012/c/c6/Metu-photos-model11.png" alt="" "> | <p><img src="https://static.igem.org/mediawiki/2012/c/c6/Metu-photos-model11.png" alt="" "> | ||
</p> | </p> | ||
| + | | ||
<p><img src="https://static.igem.org/mediawiki/2012/5/5b/Metu-photos-model12.png" alt="" "> | <p><img src="https://static.igem.org/mediawiki/2012/5/5b/Metu-photos-model12.png" alt="" "> | ||
</p> | </p> | ||
| + | | ||
<p><img src="https://static.igem.org/mediawiki/2012/a/ab/Metu-photos-model13.png" alt="" "> | <p><img src="https://static.igem.org/mediawiki/2012/a/ab/Metu-photos-model13.png" alt="" "> | ||
</p> | </p> | ||
| + | | ||
<p><img src="https://static.igem.org/mediawiki/2012/e/e7/Metu-photos-model14.png" alt="" "> | <p><img src="https://static.igem.org/mediawiki/2012/e/e7/Metu-photos-model14.png" alt="" "> | ||
</p> | </p> | ||
| + | | ||
| + | |||
</span> | </span> | ||
<h2>baslik2</h2> | <h2>baslik2</h2> | ||
Revision as of 12:03, 22 September 2012
KILL-SWITCH MODEL
The aim of kill-switch model is to predict the behavior of kill-switch mechanism in different IPTG concentrations. We first tried to model mechanism by the help of ODEs. Then we used SimBiology toolbox of MATLAB to simulate system.Introduction:
As can be seen from the diagram our circuit involves constitutive production of LacI. LacI Promoter is negatively regulated by LacI. The control mechanism of our system is inhibition of LacI by IPTG molecule. Since LacI is inhibited by IPTG its effect on LacI promoter changes with respect to IPTG concentration.
Holin and Endolysin expression is controlled by cI promoter which is negatively regulated by cI.
Our first assumption in the model is that when the free Holin molecules, which means number of Holin molecules minus number of Antiholin molecules, reach 3000[ref] cell lysis occurs.
Because cI concentration depends on the activity of LacI promoter, we can conclude that in the presence of IPTG the concentration of cI will be high and expression of Holin and Endolysin will be lowered. Due to the constitutive production of Antiholin, we can say that free Holin molecules will be lowered and chance of cell lysis to occur will drop. However, to understand the overall behaviour of system with respect to IPTG concentration we need to model and simulate the system.

Mathematical Model:
In our model, we included all processes shown below.
Equations for mRNA transcription









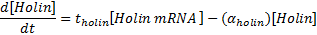


 "
"

