Team:TU Darmstadt/Modeling Homologie Modeling
From 2012.igem.org
(→TphA1) |
(→Results) |
||
| Line 132: | Line 132: | ||
===TphA1=== | ===TphA1=== | ||
[[File :TPHA1.full.png|center|700px]] | [[File :TPHA1.full.png|center|700px]] | ||
| - | [[File: TphA1_quality.png | | + | [[File: TphA1_quality.png |900px|center]] |
===TphA2=== | ===TphA2=== | ||
[[File: TPHA2.full.png |center| 700px]] | [[File: TPHA2.full.png |center| 700px]] | ||
| - | + | [[File: TphA2_quality.png |900px|center]] | |
===TphA3=== | ===TphA3=== | ||
[[File: Tph3.png.png |center|700px]] | [[File: Tph3.png.png |center|700px]] | ||
| Line 142: | Line 142: | ||
===TphB=== | ===TphB=== | ||
[[File:TPHB.full.png|center|700px]] | [[File:TPHB.full.png|center|700px]] | ||
| - | + | [[File: TphB_quality.png |900px|center]] | |
===Xyle=== | ===Xyle=== | ||
[[File: XYLE.full.png|center|700px]] | [[File: XYLE.full.png|center|700px]] | ||
Revision as of 12:37, 21 September 2012
| Homology Modeling | | Gaussian Networks | | Molecular Dynamics | | Information Theory | | Docking Simulation |
|---|
Contents |
Homology Modeling
While our proteins are functionally described in literature and during the IGEM competition, no structures are available in the protein data bank. For further work and visualizations protein structures are indispensable. We used Yasara Structure [1] to calculate 3-dimensional structures of all of our proteins for the IGEM.
Workflow
Description how our Yasara script calculates homology model[7]:
- Sequence is PSI-BLASTed against Uniprot [2]
- Calculation of a position-specific scoring matrix (PSSM) from related sequences
- Using the PSSM to search the PDB for potential modeling templates
- The Templates are ranked based on the alignment score and the structural quality[3]
- Deriving additional information’s for template and target (prediction of secondary structure, structure-based alignment correction by using SSALN scoring matrices [4]).
- A graph of the side-chain rotamer network is built, dead-end elimination is used to find an initial rotamer solution in the context of a simple repulsive energy function [5]
- The loop-network is optimized using a high amount of different orientations
- Side-chain rotamers are fine-tuned considering electrostatic and knowledge-based packing interactions as well as solvation effects.
- An unrestrained high-resolution refinement with explicit solvent molecules is run, using the latest knowledge-based force fields[6].
Application
All these steps are performed to every template used for the modeling approach. For our project we set the maximum amount of templates to 20. Every derived structure is evaluated using an average per-residue quality Z-scores. At last a hybrid model is built containing the best regions of all predictions. This procedure make prediction’s accurate and thus more realistic.
Results
PnB-Esterase 13
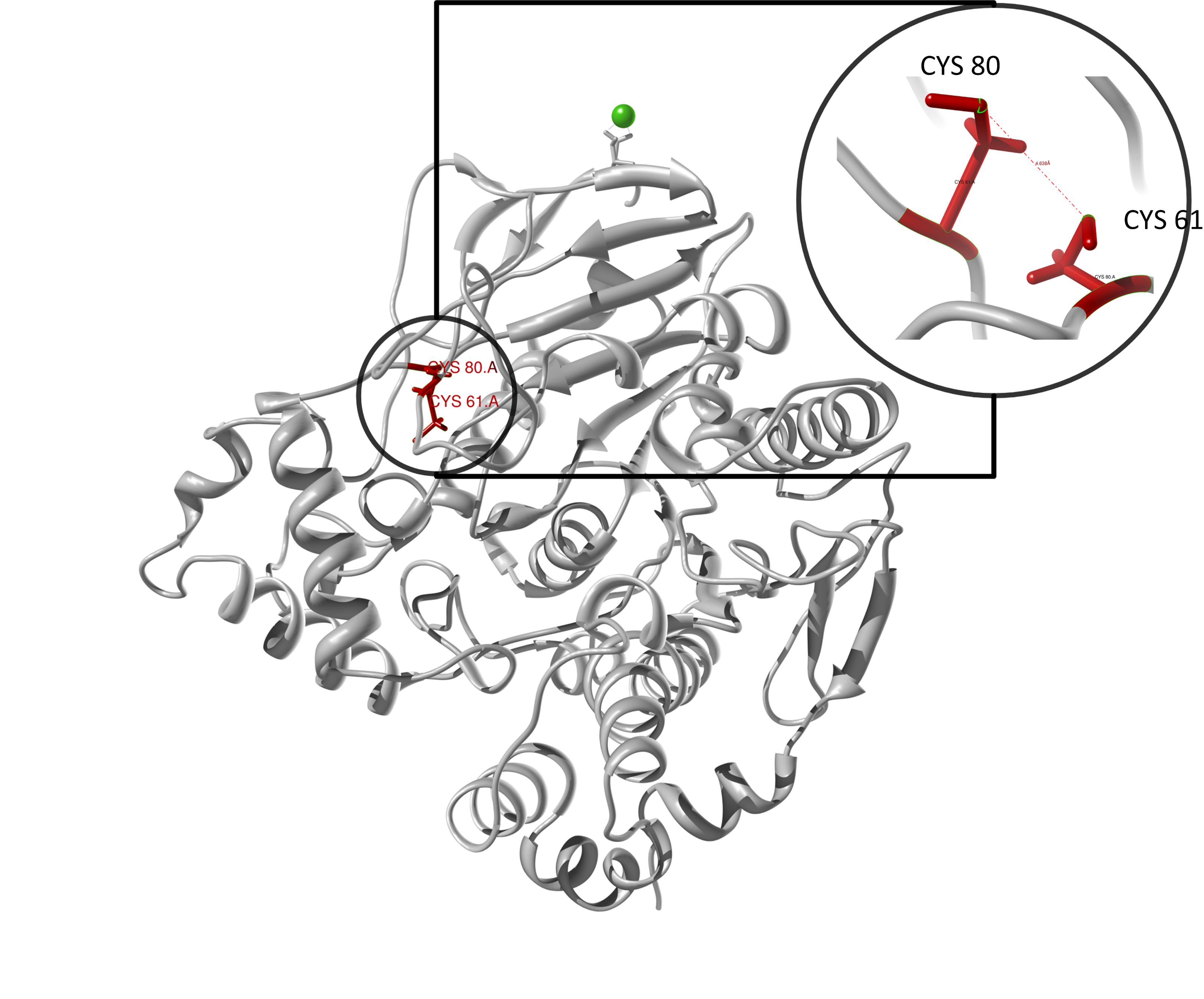
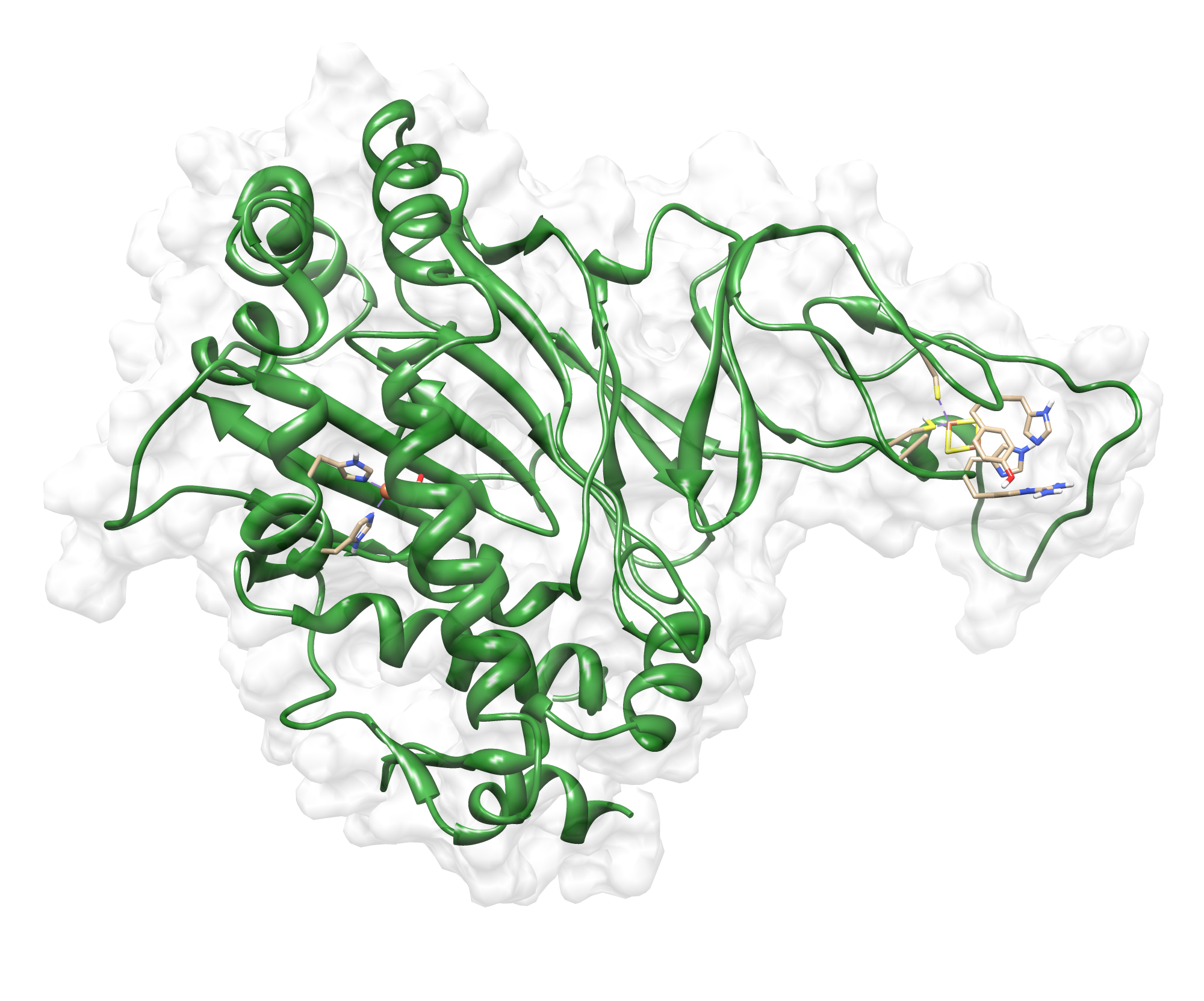
The target sequence of the PnB-Esterase 13 contains 490 residues in 1 molecule. Hence the target sequence was the only available information we identified modeling templates by running various PSI-BLAST iterations. We used 1C7J chain A, 1QE3 chain B, 1C7I chain A and 50 more! Unfortunately, our hybrid model could not be improved by copying parts from other models.
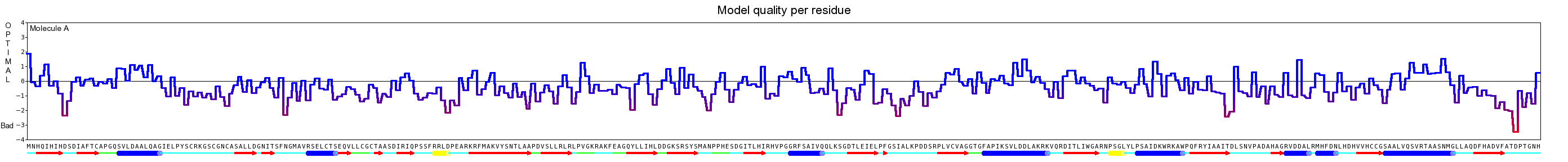
Here we show our hybrid homology model of our protein, the PnB-Esterase 13 in ribbon representation. Furthermore we illustrate the average quality z-score as a function of residue number. Nevertheless it was subjected to a final round of simulated annealing minimization in explicit solvent and obtained the following quality Z-scores:
| Check type | Quality Z-score | Comment |
|---|---|---|
| Dihedrals | -0.201 | Optimal |
| Packing 1D | -0.834 | Good |
| Packing 3D | -1.106 | Satisfactory |
| Overall | -0.661 | Good |
Hence we constructed a homology model we are able to localize a possible disulfide bond between CYS 80 and CYS 61 (highlighted in the structure above).
AroY
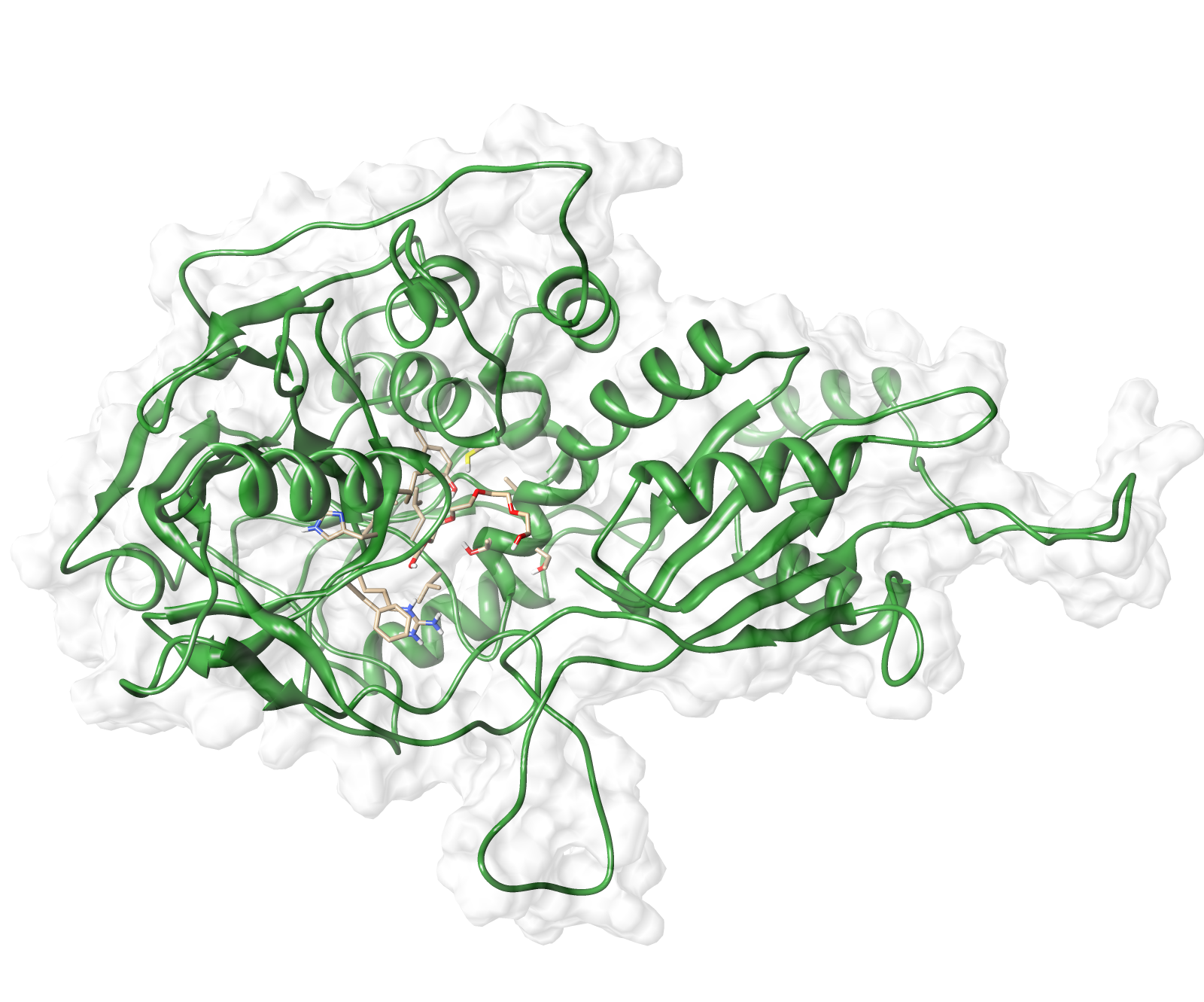
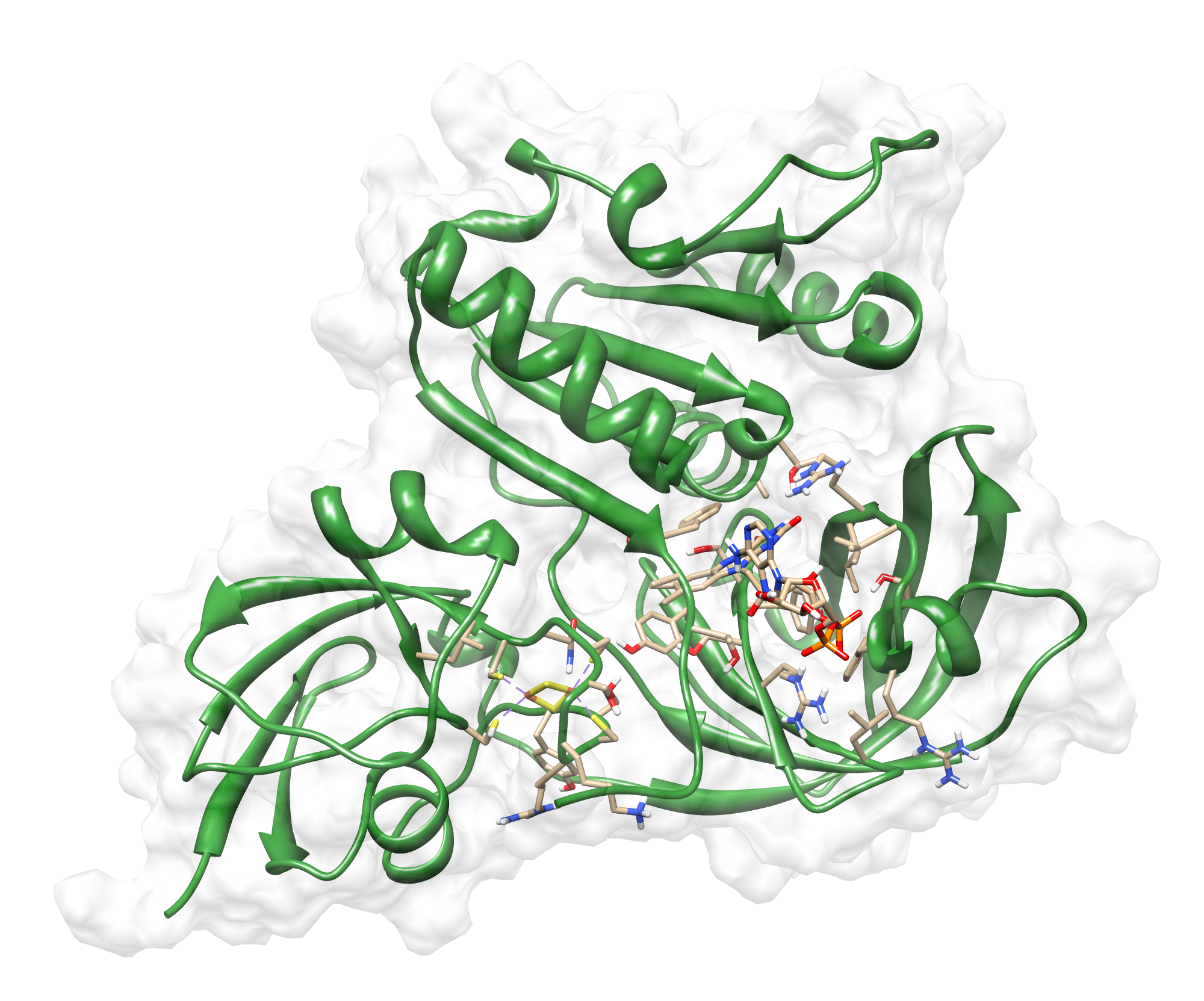
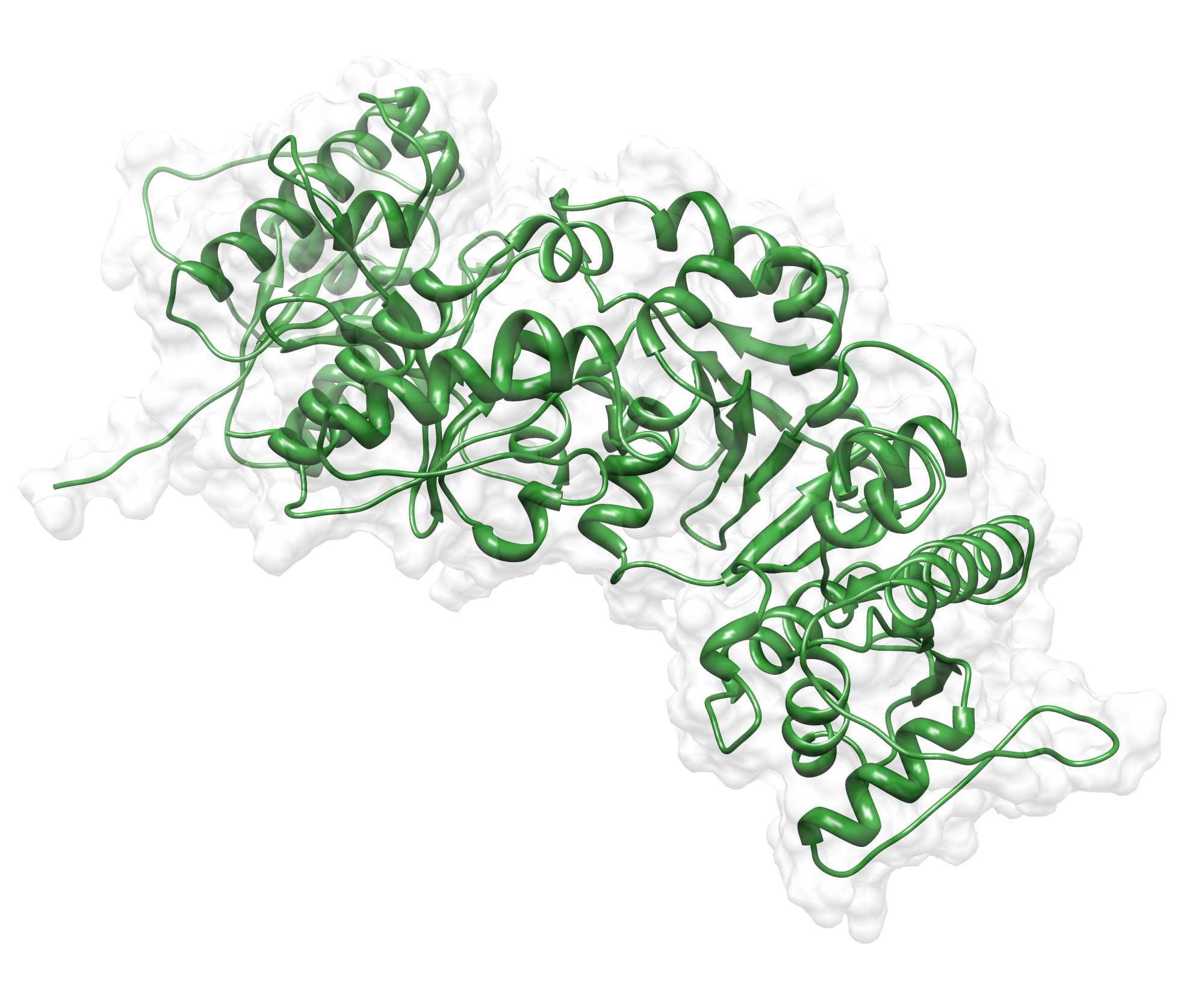
The target sequence of the AroY 13 contains 490 residues in 1 molecule. Unfortunately, we found only one model template in the protein database. We used 2IDB chain A , with an cover of 91%.
Here we show our hybrid homology model of our protein, AroY in ribbon representation with transparent surface. Furthermore we illustrate the average quality z-score as a function of residue number. The total results are listed in the tabular below.
| Check type | Quality Z-score | Comment |
|---|---|---|
| Dihedrals | 0.454 | Optimal |
| Packing 1D | -2.814 | Poor |
| Packing 3D | -2.382 | Poor |
| Overall | -2.139 | Poor |
Since the overall quality Z-scores was ranked poor by Yasara we refined the bonding-network and applied a simulated annealing energy minimization.
TphA1
TphA2
TphA3
TphB
Xyle
References
[1] E. Krieger, G. Koraimann, and G. Vriend, “Increasing the precision of comparative models with YASARA NOVA--a self-parameterizing force field.,” Proteins, vol. 47, no. 3, pp. 393–402, 2002.
[2] S. F. Altschul, T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman, “Gapped BLAST and PSI-BLAST: a new generation of protein database search programs.,” Nucleic Acids Res, vol. 25, no. 17, pp. 3389–3402, Sep. 1997.
[3] R. W. Hooft, G. Vriend, C. Sander, and E. E. Abola, “Errors in protein structures.,” Nature, vol. 381, no. 6580. Nature Publishing Group, p. 272, 1996.
[4] D. T. Jones, “Protein secondary structure prediction based on position-specific scoring matrices,” Journal of Molecular Biology, vol. 292, no. 2, pp. 195–202, 1999.
[5] A. A. Canutescu, A. A. Shelenkov, and R. L. Dunbrack, “A graph-theory algorithm for rapid protein side-chain prediction.,” Protein Science, vol. 12, no. 9, pp. 2001–2014, 2003.
[6] E. Krieger, K. Joo, J. Lee, J. Lee, S. Raman, J. Thompson, M. Tyka, D. Baker, and K. Karplus, “Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8.,” Proteins, vol. 77 Suppl 9, no. June, pp. 114–122, 2009.
[7] http://www.yasara.org/homologymodeling.htm
 "
"