Team:USTC-China/futurework
From 2012.igem.org
| Line 108: | Line 108: | ||
<h2><a name="2.1">Plasmid Keeping System</a></h3> | <h2><a name="2.1">Plasmid Keeping System</a></h3> | ||
| - | <div class="imgholder1" align="left" style="float:left;width: | + | <div class="imgholder1" align="left" style="float:left;width:100%;height:auto;"> |
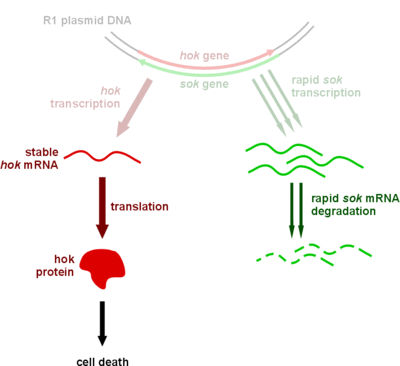
<a href="https://static.igem.org/mediawiki/2012/3/31/Mechanism_of_the_hoksok_system_in_bacteria_in_the_presence_of_R1_plasmid_DNA.gif"><img src="https://static.igem.org/mediawiki/2012/3/31/Mechanism_of_the_hoksok_system_in_bacteria_in_the_presence_of_R1_plasmid_DNA.gif" alt="Mechanism_of_the_hoksok_system_in_bacteria_in_the_presence_of_R1_plasmid_DNA" style="clear:both;width:289px;height:254px;"></a><br><small align="center">Mechanism of the hok/sok system in bacteria in the presence of R1 plasmid DNA</small> | <a href="https://static.igem.org/mediawiki/2012/3/31/Mechanism_of_the_hoksok_system_in_bacteria_in_the_presence_of_R1_plasmid_DNA.gif"><img src="https://static.igem.org/mediawiki/2012/3/31/Mechanism_of_the_hoksok_system_in_bacteria_in_the_presence_of_R1_plasmid_DNA.gif" alt="Mechanism_of_the_hoksok_system_in_bacteria_in_the_presence_of_R1_plasmid_DNA" style="clear:both;width:289px;height:254px;"></a><br><small align="center">Mechanism of the hok/sok system in bacteria in the presence of R1 plasmid DNA</small> | ||
| - | |||
| - | |||
<a href="https://static.igem.org/mediawiki/2012/9/99/Mechanism_of_the_hoksok_system_in_bacteria_in_the_absence_of_R1_plasmid_DNA.gif"><img src="https://static.igem.org/mediawiki/2012/9/99/Mechanism_of_the_hoksok_system_in_bacteria_in_the_absence_of_R1_plasmid_DNA.gif" alt="Mechanism_of_the_hoksok_system_in_bacteria_in_the_absence_of_R1_plasmid_DNA" style="clear:both;width:289px;height:254px;"></a><br><small align="center">Mechanism of the hok/sok system in bacteria in the absence of R1 plasmid DNA</small> | <a href="https://static.igem.org/mediawiki/2012/9/99/Mechanism_of_the_hoksok_system_in_bacteria_in_the_absence_of_R1_plasmid_DNA.gif"><img src="https://static.igem.org/mediawiki/2012/9/99/Mechanism_of_the_hoksok_system_in_bacteria_in_the_absence_of_R1_plasmid_DNA.gif" alt="Mechanism_of_the_hoksok_system_in_bacteria_in_the_absence_of_R1_plasmid_DNA" style="clear:both;width:289px;height:254px;"></a><br><small align="center">Mechanism of the hok/sok system in bacteria in the absence of R1 plasmid DNA</small> | ||
</div> | </div> | ||
Revision as of 15:21, 18 September 2012
Future Work
Future Directions
Though the iGEM 2012 is coming to an end,we plan to do further research to make our research accomplished and endue it with new functions. In the future, with enough time and abundant knowledge, these meaningful designs will be realized.
Plasmid Keeping System
We plan to construct a plasmid keeping system to avoid plasmid loss. In phage-free situations, the bacteria is likely to lose the anti-phage plasmid, therefore a plasmid keeping system is necessary. We choose the hok/sok system to achieve this. It belongs to type I toxin-antitoxin system. The cell will die if the cell lost the plasmid containing the system. Thus, only those cells contain survives. Besides, the type I toxin-antitoxin system is an RNA based system, therefore it won't spend too much energy.
Quorum Sensing
Actually, at the primary plan of our project, we thought that when the suicide gene works, the host would use quorum sensing to alarm the bacterial around and make them prepare the defense. We don't hope the bacterial spend a lot of energy on the defense when there is no phage is invading. To this end, we design that the defense action will be taken only when the bacterial receive the quorum sensing signal, which is sent by the host. The reason for giving up this feature is that we haven't found any effective defense method. The defense method we adopted is blocking the phage’s binding site on the membrane of bacterial. We learned that a sugar transporter called LamB (the structure is showed below) may be the phage’s binding site and we planned to use aptamer to block the interaction between the LamB and the lambda phage.
But unfortunately, there are too many difficulties we have to face. For example, we can't make the aptamer go through the inner membrane of E.coli to get the target which is on the outer membrane. What’s more, the sugar transporter LamB on the outer membrane is too complex for us to choose the binding site of the aptamer, which across the membrane for 18 times (see the proposed model for the folding of LamB in the outer membrane.).
Application To Fermentation Industry
We plan to apply this method to yeast fermentation industry.
Cure For Viral Diseases
If possible, we will make some medical research with this method, because this anti-virus method is universal. It will be a possible way on therapies against viral diseases.
 "
"