Team:Freiburg/Project/Overview
From 2012.igem.org
| Line 65: | Line 65: | ||
|[[Image:Extension3.png|600px|no frame]] | |[[Image:Extension3.png|600px|no frame]] | ||
|} | |} | ||
| + | |||
| + | |||
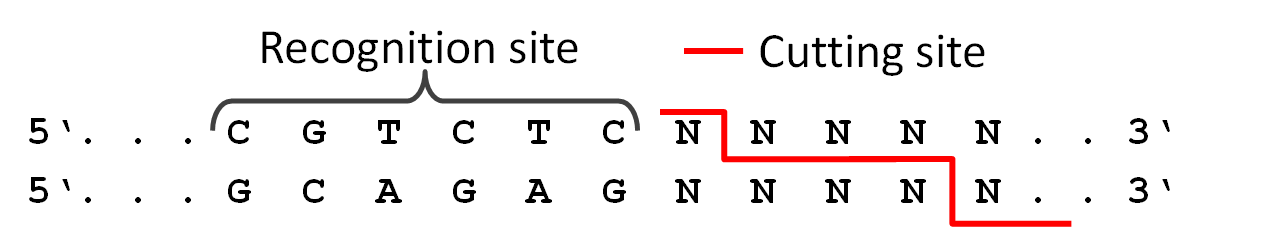
| + | Because we didnt intend to require six different restriction enzyms in the final pcr to produce a twelve direpeat TAL we used a technic called golden gate cloning. The technic uses the type two restriction enzyme BsmB1 and its ability to cut DNA slightly downstream of the recognition site. With this it is possible for us to create different sticky ends with using just one enzyme. Therefore we are able to ligate six different parts in the right order in one pcr step. | ||
| + | |||
| + | |||
| + | {|align="center" | ||
| + | |[[Image:bsmb1.png|500px|no frame]] | ||
| + | |} | ||
| + | |||
| Line 76: | Line 85: | ||
| + | ==<span style="color:#2244AA"> Part 2: The TAL vektor == | ||
| + | |||
| + | |||
| + | The next step of our project was the creation of a working vektor in which we could ligate our TAL protein for tranformation or transfection. | ||
<!--- The Mission, Experiments ---> | <!--- The Mission, Experiments ---> | ||
Revision as of 16:21, 9 September 2012
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modeling | Notebook | Safety | Attributions |
|---|
| Building a Toolkit | Using the Toolkit | Experiments |
|---|
Building a Toolkit
Part 1: The 96 direpeats
What we are planned to do:
|
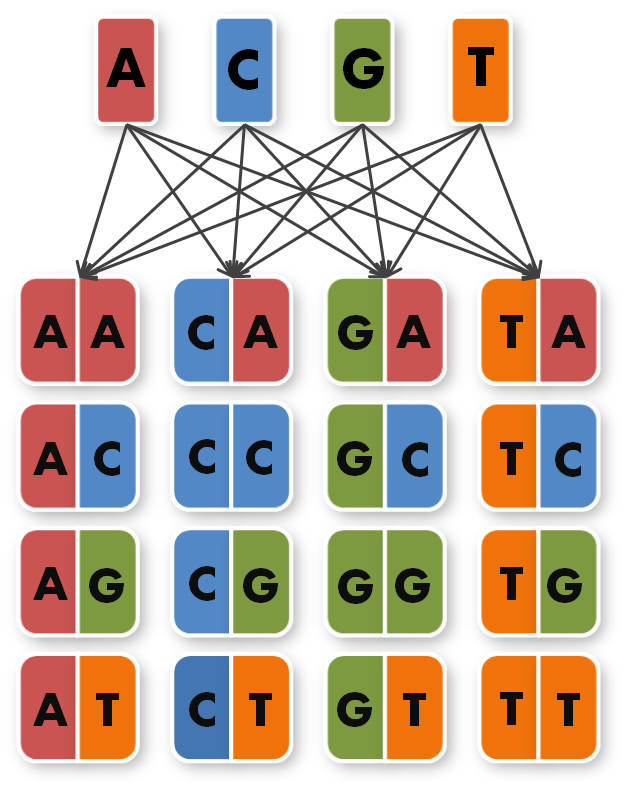
We took the sequences of the four already known TAL Repeats A,C,G,T and combined them to 16 new, so called direpeat sequences. These 16 direpeats were ordered as gene synthesis products.
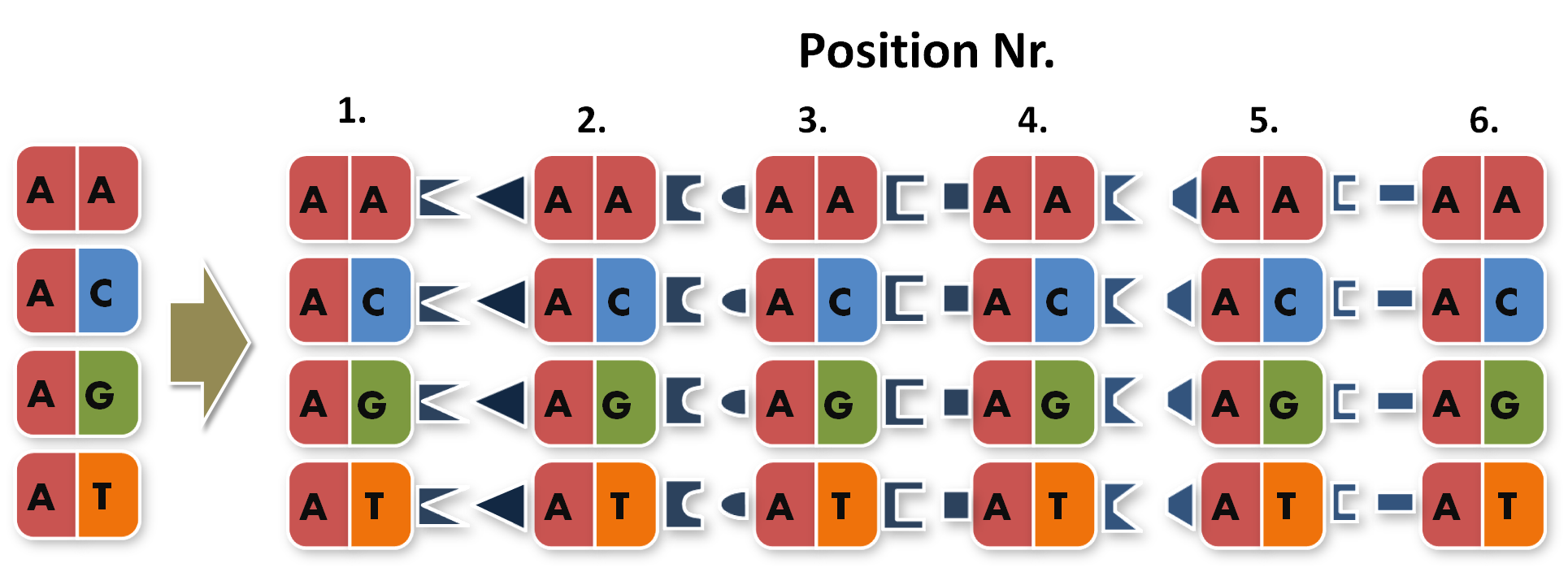
Now the real work began, to start building TAL Proteins we needed to expand our 16 direpeats a second time. Each of the 16 direpeats needed to be split into six versions, one version for every place of our final six direpeat respectevly twelve monorepeat TAL Protein. Because we didnt wanted to buy six times 16 different synthesised direpeats we came up with a plan to produce them by ourself. We created six primer pairs each primer with a common part matching everyone of the direpeats and an unique overhang to extend the direpeat it binds to.
|
Because we didnt intend to require six different restriction enzyms in the final pcr to produce a twelve direpeat TAL we used a technic called golden gate cloning. The technic uses the type two restriction enzyme BsmB1 and its ability to cut DNA slightly downstream of the recognition site. With this it is possible for us to create different sticky ends with using just one enzyme. Therefore we are able to ligate six different parts in the right order in one pcr step.
|
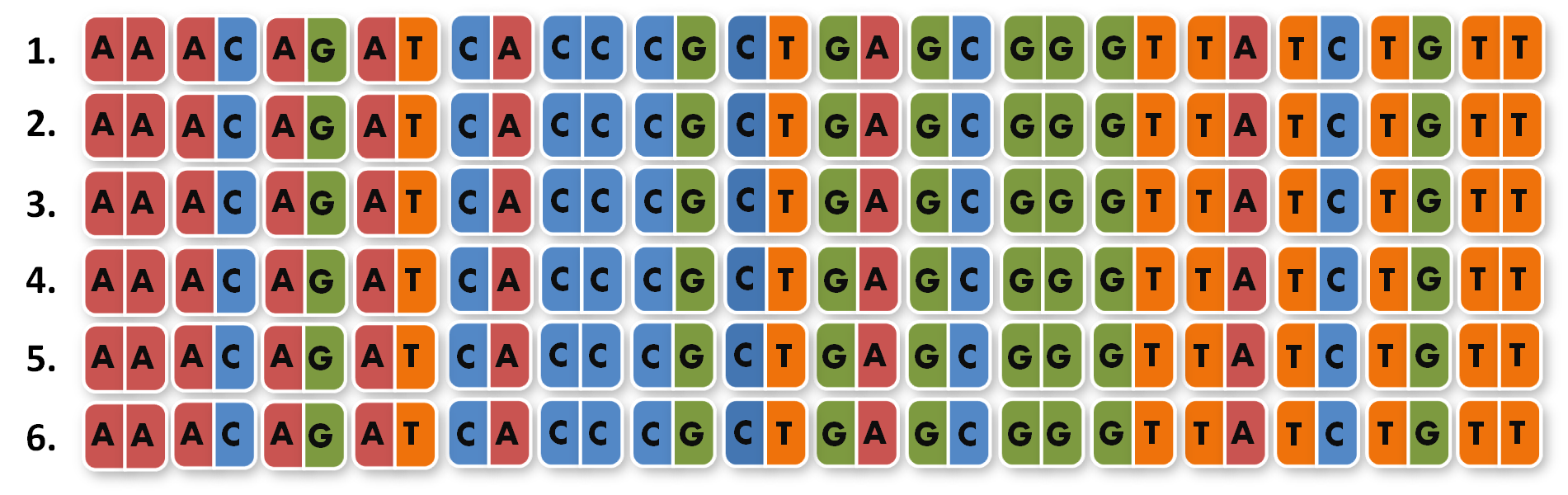
After finshing these 96 different extension pcr's we had to ligate the products into the orignial igem biobrick vektor and finally got our full library of 96 unique direpeats
|
Part 2: The TAL vektor
The next step of our project was the creation of a working vektor in which we could ligate our TAL protein for tranformation or transfection.
 "
"
