Team:USTC-China/results
From 2012.igem.org
(Difference between revisions)
| Line 98: | Line 98: | ||
<p>Calculate the natural logarithm of both side of the equation,we can get:</p> | <p>Calculate the natural logarithm of both side of the equation,we can get:</p> | ||
<p>Obviously,the variable lnx is</p> | <p>Obviously,the variable lnx is</p> | ||
| + | <div class="imgholder1" align="left" style="float:right;width:172px;height:auto;"> | ||
| + | <a href="https://static.igem.org/mediawiki/2012/4/48/Formulaustc1.jpg"><img src="https://static.igem.org/mediawiki/2012/4/48/Formulaustc1.jpg" alt="Curve" style="clear:both;width:172;height:50px;"></a><br></div> | ||
<p>linear to the variable t.And the slope is μm.</p> | <p>linear to the variable t.And the slope is μm.</p> | ||
<p>We calculate the natural logarithm of OD600 data we get and do linear fit to the data.Then we get the slope which is the natural growth rate μm.</p> | <p>We calculate the natural logarithm of OD600 data we get and do linear fit to the data.Then we get the slope which is the natural growth rate μm.</p> | ||
| Line 104: | Line 106: | ||
| - | <div class="imgholder1" align="left" style=" | + | <div class="imgholder1" align="left" style="width:700px;height:auto;"> |
<a href="https://static.igem.org/mediawiki/2012/b/b1/Curve.jpg"><img src="https://static.igem.org/mediawiki/2012/b/b1/Curve.jpg" alt="Curve" style="clear:both;width:750;height:300px;"></a><br><small align="center">Figure 1 Growth curve of the two groups. (a)experiment group prm-anticro-prm-lysis (b)control group croGFP</small></div> | <a href="https://static.igem.org/mediawiki/2012/b/b1/Curve.jpg"><img src="https://static.igem.org/mediawiki/2012/b/b1/Curve.jpg" alt="Curve" style="clear:both;width:750;height:300px;"></a><br><small align="center">Figure 1 Growth curve of the two groups. (a)experiment group prm-anticro-prm-lysis (b)control group croGFP</small></div> | ||
| - | <div class="imgholder1" align="left" style=" | + | <div class="imgholder1" align="left" style="width:700px;height:auto;"> |
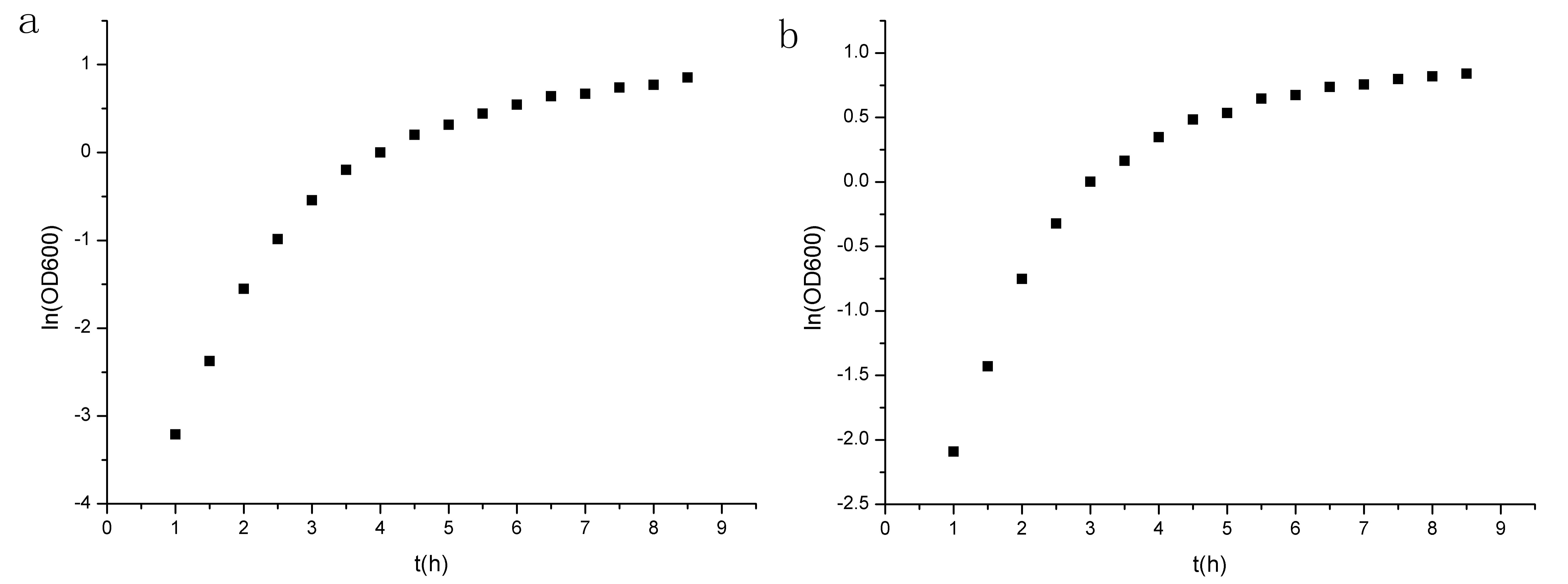
<a href="https://static.igem.org/mediawiki/2012/9/96/Points.jpg"><img src="https://static.igem.org/mediawiki/2012/9/96/Points.jpg" alt="Points" style="clear:both;width:750;height:300px;"></a><br><small align="center">Figure 2 Points of ln(OD600) to t. (a)experiment group prm-anticro-prm-lysis (b)control group croGFP </small></div> | <a href="https://static.igem.org/mediawiki/2012/9/96/Points.jpg"><img src="https://static.igem.org/mediawiki/2012/9/96/Points.jpg" alt="Points" style="clear:both;width:750;height:300px;"></a><br><small align="center">Figure 2 Points of ln(OD600) to t. (a)experiment group prm-anticro-prm-lysis (b)control group croGFP </small></div> | ||
<p>We select the four data points and do linear fit.Then we can get μm,exp(experimental group) and μm,ctrl(control group).</p> | <p>We select the four data points and do linear fit.Then we can get μm,exp(experimental group) and μm,ctrl(control group).</p> | ||
| - | <div class="imgholder1" align="left" style=" | + | <div class="imgholder1" align="left" style="width:700px;height:auto;"> |
<a href="https://static.igem.org/mediawiki/2012/0/0a/Fitlysis01.png"><img src="https://static.igem.org/mediawiki/2012/0/0a/Fitlysis01.png" alt="Fitlysis01" style="clear:both;width:750;height:300px;"></a><br><small align="center">Figure 3 Linear fit of ln(OD600) to t. (a)experiment group prm-anticro-prm-lysis (b)control group croGFP</small></div> | <a href="https://static.igem.org/mediawiki/2012/0/0a/Fitlysis01.png"><img src="https://static.igem.org/mediawiki/2012/0/0a/Fitlysis01.png" alt="Fitlysis01" style="clear:both;width:750;height:300px;"></a><br><small align="center">Figure 3 Linear fit of ln(OD600) to t. (a)experiment group prm-anticro-prm-lysis (b)control group croGFP</small></div> | ||
<table border="1" style="text-align:center;"> | <table border="1" style="text-align:center;"> | ||
Revision as of 15:25, 23 September 2012
RESULTS
 "
"