Team:UC Chile2/Cyanolux/Project
From 2012.igem.org
| Line 63: | Line 63: | ||
<br /> | <br /> | ||
| - | + | <html> | |
| + | <img src="https://static.igem.org/mediawiki/2012/5/56/Alagain_uc_chile.jpg" width="300" align="left" style ="margin-right:15px"></html> | ||
<p>LuxAB genes have been widely used as reporters dependent on the addition of n-decanal to the culture | <p>LuxAB genes have been widely used as reporters dependent on the addition of n-decanal to the culture | ||
media and in 2010, the Cambridge iGEM team engineered LuxABCDEG to an <i>E. coli</i>-optimized biobrick | media and in 2010, the Cambridge iGEM team engineered LuxABCDEG to an <i>E. coli</i>-optimized biobrick | ||
| Line 72: | Line 73: | ||
produced by this pathway is much more visually appealing than other systems from the registry (i.e XFPs), | produced by this pathway is much more visually appealing than other systems from the registry (i.e XFPs), | ||
moreover, the light production doesn´t depend on a single peptide but on a whole pathway involving several genes, which makes it much more tunable, for instance, decoupling in time the substrate recovery from the luciferase reaction itself.</p> | moreover, the light production doesn´t depend on a single peptide but on a whole pathway involving several genes, which makes it much more tunable, for instance, decoupling in time the substrate recovery from the luciferase reaction itself.</p> | ||
| + | |||
| + | |||
Revision as of 18:45, 23 September 2012

Contents |
Motivational drive
Natural cycles have always fascinated mankind, probably due to the mysterious mechanisms involved in them and the power they exert in our everyday life. Since the dawn of synthetic biology, engineering oscillatory systems has been a recurrent topic, being Ellowitz's represillator a classical example. Nevertheless, to date no iGEM team has accomplished the implementation of a robust oscillatory system. That will be our challenge for this year's iGEM project.
To reach our goal we designed a synthethic circuit that links to the endogenous circadian rhythm of Synechocystis PCC6803. As a proof of concept we are going to engineer the first light-rechargeable biological lamp: Synechocystis PCC 6803 cells that emit light only by night while recovering and producing the substrates in the day. We strongly believe this will serve as an enabling tool to any project requiring time control over a biological behaviour independently of the user's input.
Furthermore, the characterization of this chassis is a fundamental step to explore new systems with minimal inputs to replace E. coli, for example, in the biotechnological industry in order to achieve greener processes.
Background
Synechocystis PCC 6803

Cyanobacteria are prokaryotic photoautotrophs and they are believed to be the only group of organisms to evolve oxygenic photosynthesis about 2.4 billion years ago 1. Although this biochemical breakthrough can’t be understated, several cyanobacteria species also play a crucial role in the planet nitrogen cycle as marine diazotrophs 2. They are found almost in every environment in earth´s surface and interestingly, they are the only prokaryote known to have circadian rhythms, probably accounting for their photosyntethic lifestyle 3.
Synechocystis PCC6803 is a gram negative non-filamentous cyanobacteria and it was the third prokaryote and the first photoautotroph whose genome was sequenced. Consequently, is has become a model organism as its genetic background has been widely studied.
Given the reasons mentioned above, it is of no surprise that Synechocystis PCC6803 (among other cyanobacteria) has been extensively used for biotechnological applications and proposed as the “green E.coli”4.
With the dawn of Synthetic Biology, research has made use of Synechocystis for commodity chemicals production and detection of water soluble pollutants among other applications5,6.
While aware of these practical applications, we (UC_Chile) are particulary interested in the well characterized circadian behavior of this chassis 7 and its implications. There are several genes known to oscillate in a daily basis, most of them related to respiration, photosynthesis and energy metabolism 8, and it has been shown that reporter systems using these promoters show a similar expression pattern 9.
Lastly, there are a lot of biobricks designed especially for Synechocystis or from its genome´s sequence by previous iGEM teams (references) but sadly no one has ever characterized them in this chassis and the registry lacks a set of tools for its transformation with standard biological parts. Moreover, to our knowledge, no iGEM team has ever accomplished a successful direct Synechocystis transformation with naked DNA.
Lux Operon
The lux operon is a group of genes that are responsible for density-dependent bioluminescent behavior in various prokariotic organisms such as Vibrio fischeri and Photorabdus luminescens. In V. fischeri, the operon is composed of 8 genes: LuxA and LuxB encode for the monomers of a heterodimeric luciferase; LuxC, LuxD and LuxE code for fatty acid reductases enzymes and LuxR and LuxI are responsible for the regulation of the whole operon.
Lastly LuxG is believed to act as a FMNH2 dependent FADH reductase, although luminescence is barely affected in its absence. The n-decanal ( n= 9 to 14) substrate oxidization to n-decanoic acid by the LuxAB heterodimer is coupled with the reduction of FMNH to FMNH2 and the releasing of oxygen and x photons of light at x wavelength.
The carboxylic group of the product is then reduced to aldehyde by CDE proteins allowing the reaction to start over.

LuxAB genes have been widely used as reporters dependent on the addition of n-decanal to the culture media and in 2010, the Cambridge iGEM team engineered LuxABCDEG to an E. coli-optimized biobrick format, uncoupling it from the LuxR and LuxI regulation.
As a team we decided to work with this operon for a number of reasons, first of all, the luminescence produced by this pathway is much more visually appealing than other systems from the registry (i.e XFPs), moreover, the light production doesn´t depend on a single peptide but on a whole pathway involving several genes, which makes it much more tunable, for instance, decoupling in time the substrate recovery from the luciferase reaction itself.
Experimental Strategy
We have devised different strategies to achieve bioluminescence controlled under circadian rhythms. Here we describe the strategies used for building the constructs to reach our goals.
Splitting the Lux operon and choosing promoters
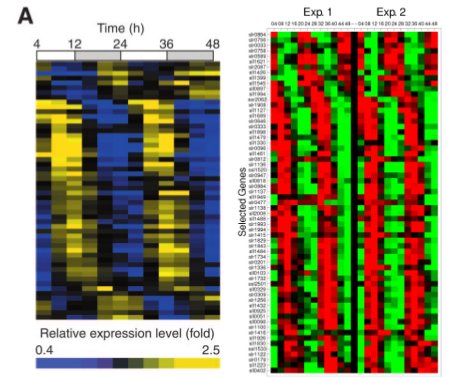
We decided that we would separate LuxAB (the luciferase part of the operon) and LuxCDEG (the substrate producing enzymes LuxCDE with LuxG the FMNH2/FMN reducing enzyme) to allow phase-dependent expression of the parts. Using specific promoters of Synechocystis PCC. 6803 we can have fine-tunning of the production of bioluminescence. Recent work on global gene expression in Synechocystis aided on finding adecuate promoters 7, 8 . (Images at right from cited papers.)
To try our approach, we selected various promoters which could serve the purpose. Our rational for selecting candidate promoters involved amplitude of oscillation, peak activity, hour, absence of restriction sites, predicted strength of promoter according to the role of the gene and reproducibility between experiments (based on the literature available). We looked for promoters which would have peak expression nearby dusk hours and that were slightly out of phase to optimize production of bioluminescence according to our mathematical models (LINK OVER HERE!). We prioritized promoters from genes that would be involved in central energetic metabolism as we believe that their expression would be most robust and reliable.
We choose the transaldolase promoter (specific name here and code in Synechocystis Genome) to direct the expression of the LuxAB genes and we found a couple of other promoters which filled the other requirements from above. Pcaa3 (NAME HERE AND DESCRIPTION OF ENDOGENOUS ACTIVITY) and PsigE (NAME HERE AND DESCRIPTION OF ENDOGENOUS ACTIVITY), the former being already in Biobrick format (courtesy from the Utah team iGEM 2010).
Building constructs
When confronted with the different available strategies to express the genes from the Lux operon in Synechocystis, we concluded that the one that best suits our need is by using integration plasmids. The reason for this is that the available plasmids that replicate in Synechocystis are very large (8 Kb) without even considering the genes we need to include in the constructs (that would sum up to a final 16 Kb aproximately). Such a large plasmid would prove very difficult to handle through molecular biology techniques, let alone transform Synechocystis.
Using integration plasmids also proposes an additional advantage, that is to produce successive integrations which allow accumulation of desirable elements in its genome. Integration in Synechocystis is undergone through double recombination of homologous DNA which also allows interruption of genes if wanted. In our case we have designed our system to produce suceptibility to copper as a biosafety measure to have further control over our recombinant Synechocystis.
We have designed two constructs that have different recombination locations in the Synechocystis chromosome. We have named them according to what Utah iGEM team from 2010 proposed for naming conventions:
pSB1C3_IntK
BBa_K743006
This constructs is an integrative plasmid which targets neutral recombination sites (slr0370 and sll0337). We selected this locus because it has been extensively used in the literature (CAPAZ EXAGERE?) and it shown to have no deleterious effects on Synechocystis viability. We selected Kanamycin resistance as our transformation marker. [PUT LINK TO CONSTRUCT HERE].
pSB1C3_IntS
We designed another construct that besides serving as a double recombination plasmid it makes Synechocystis susceptible to copper concentrations higher than X uM 10. We have designed this construct to interrupt the CopS gene as a biosafety measure to avoid the possibility of having a leakage of recombinant DNA to the environment. This plasmid has Spectynomycin resistance as the transformation marker. [PUT LINK TO CONSTRUCT HERE]
pSB1A3_IntC (Utah 2010 iGEM Team integration plasmid)
Alternatively, we designed our LuxCDEG contructs for the Utah 2010 iGEM Team plasmid backbone . pSB1A2_IntC.
References
(1) Evolution of photosynthesis. Hohmann-Marriott MF, Blankenship. Annual Review of Plant Biology Vol. 62: 515-548
(2) Nitrogen fixation by marine cyanobacteria. Jonathan P. Zehr. Trends in microbiology, Volume 19, Issue 4, April 2011, Pages 162–17
(3) Carl Hirschie Johnson and Susan S. Golden. CIRCADIAN PROGRAMS IN CYANOBACTERIA: Adaptiveness and Mechanism. Annual Review of Microbiology, Vol. 53: 389-409
(4) Ducat, D. C., Way, J. C., & Silver, P. a. (2011). Engineering cyanobacteria to generate high-value products. Trends in biotechnology, 29(2), 95-103.
(5) Huang, H.-H., Camsund, D., Lindblad, P., & Heidorn, T. (2010). Design and characterization of molecular tools for a Synthetic Biology approach towards developing cyanobacterial biotechnology. Nucleic acids research, 38(8), 2577-93
(6) Peca, L., Kós, P. B., Máté, Z., Farsang, A., & Vass, I. (2008). Construction of bioluminescent cyanobacterial reporter strains for detection of nickel, cobalt and zinc. FEMS microbiology letters, 289(2), 258-64.
(7) Kucho, K.-ichi, Okamoto, K., Tsuchiya, Y., Nomura, S., Nango, M., Kanehisa, M., Ishiura, M., et al. (2005). Global Analysis of Circadian Expression in the Cyanobacterium Synechocystis sp . Global Analysis of Circadian Expression in the Cyanobacterium. Society.
(8) Layana, C., & Diambra, L. (2011). Time-course analysis of cyanobacterium transcriptome: detecting oscillatory genes. PloS one, 6(10), e26291.
(9) Kunert, a, Hagemann, M., & Erdmann, N. (2000). Construction of promoter probe vectors for Synechocystis sp. PCC 6803 using the light-emitting reporter systems Gfp and LuxAB. Journal of microbiological methods, 41(3), 185-94.
(10)Giner-Lamia, J., Lopez-Maury, L., Reyes, J. C., & Florencio, F. J. (2012). The CopRS two-component system is responsible for resistance to copper in the cyanobacterium Synechocystis sp. PCC 6803. Plant physiology, 159(August), 1806-1818.
 "
"