Team:UC Chile/Results/Int C
From 2012.igem.org
(Difference between revisions)
| Line 2: | Line 2: | ||
| - | < | + | <h1>Int_C transformation</h1> |
[[File:Int_C-Failagain.JPG| 600px| center]] | [[File:Int_C-Failagain.JPG| 600px| center]] | ||
| Line 8: | Line 8: | ||
<br> | <br> | ||
| - | < | + | <h1>pSB1A3_IntC (From 2010 Utah's iGEM team)</h1> |
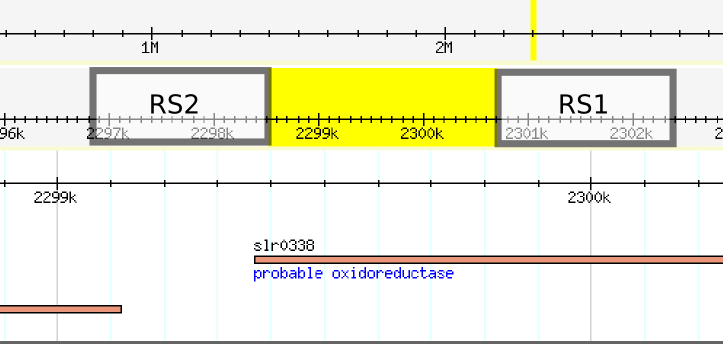
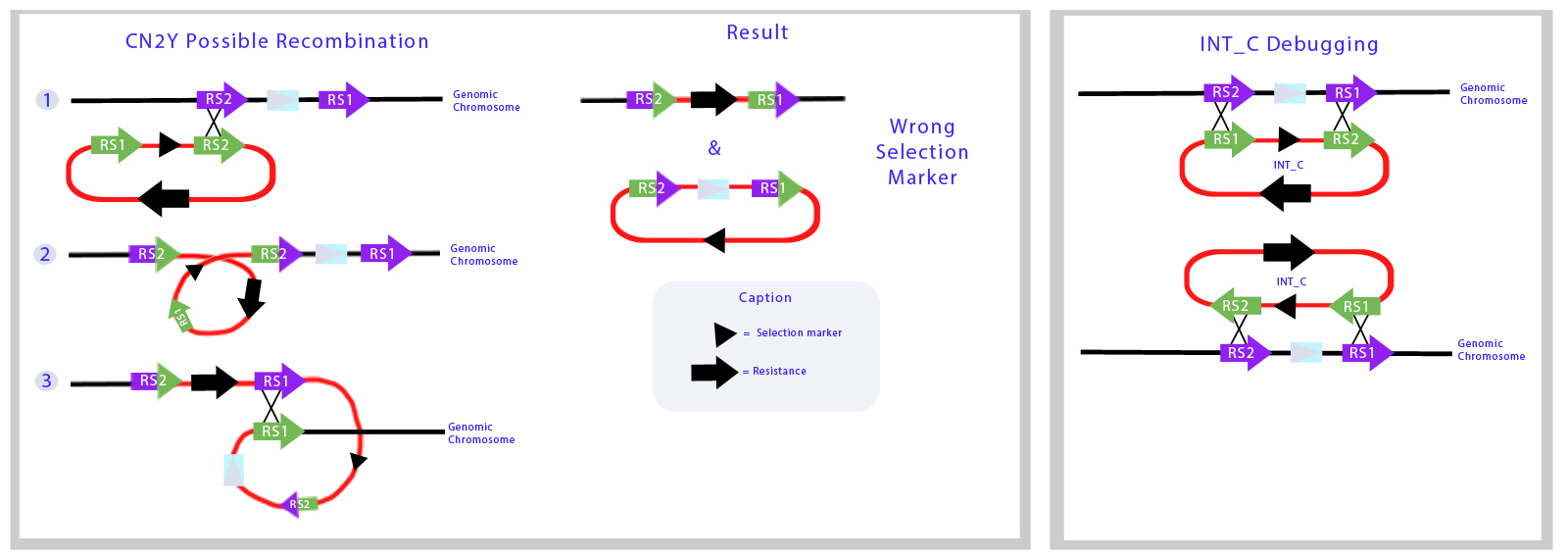
We decided to use this plasmid to place the LuxCDEG part of the Lux operon in this plasmid, however after various attempts to transform Synechocystis without success we reconsidered (see below SOMEWHERE!!! [[]]) due to problems regarding the design of this plasmid backbone. After analyzing the sequence of the vector (CODE HERE) we blasted the sequences of the recombination sites and we found out that position of the sequences of each recombination site are swapped, leaving the backbone part of the plasmid to be integrated into the genome. Furthermore, the recombination sites are separated by aproximately 2.2 kilobases, knocking 1 gene. | We decided to use this plasmid to place the LuxCDEG part of the Lux operon in this plasmid, however after various attempts to transform Synechocystis without success we reconsidered (see below SOMEWHERE!!! [[]]) due to problems regarding the design of this plasmid backbone. After analyzing the sequence of the vector (CODE HERE) we blasted the sequences of the recombination sites and we found out that position of the sequences of each recombination site are swapped, leaving the backbone part of the plasmid to be integrated into the genome. Furthermore, the recombination sites are separated by aproximately 2.2 kilobases, knocking 1 gene. | ||
Revision as of 07:23, 26 September 2012
 "
"