Team:UC Chile/Results/Int C
From 2012.igem.org
(Difference between revisions)
(Created page with "{{UC_Chile4}} <h2>pSB1A3_IntC (From 2010 Utah's iGEM team)</h2> We decided to use this plasmid to place the LuxCDEG part of the Lux operon in this plasmid, however after various...") |
|||
| (8 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{UC_Chile4}} | {{UC_Chile4}} | ||
| - | |||
| - | |||
| - | + | <h1>Debugging pSB1A3_IntC</h1> | |
| + | <br /> | ||
| + | |||
| + | After repeatedly having failed transforming Synechocystis PCC. 6803 (3 times) with pSB1A3_IntC , we sought to analyze the sequence of the plasmid we acquired from the 2010 Utah iGEM team by sequencing the recombination sites. | ||
| + | |||
| + | <div style="width: 600px; padding: 10px; margin-right: auto; margin-left: auto"> | ||
| + | [[File:Int_C-Failagain.JPG| 600px| center]] | ||
| + | Image from the 3rd failed transformation of Synechocystis PCC 6803 with the 2010 Utah iGEM team [http://partsregistry.org/Part:BBa_K390200 pSB1A3_IntC ] (BBa_K390200) plasmid. Although you might see a green stain on the plate, this plate has been incubating for over 4 weeks and still there are no colonies. | ||
| + | </div> | ||
| + | |||
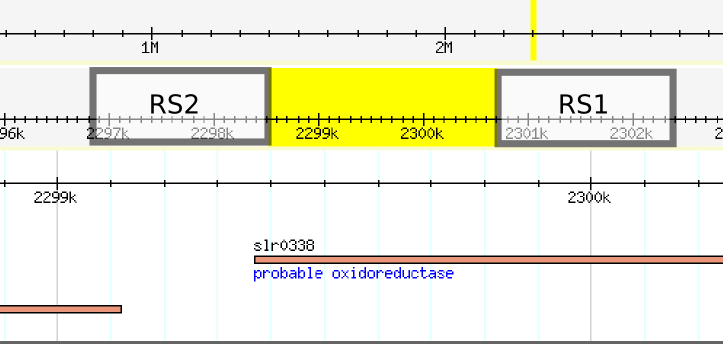
| + | While we did not find any errors in the sequence, we noticed when blasting each separate recombination site, that the position of the recombination sites differed from the ones reported on their page. The position of the sequences of each recombination site are swapped, leaving the backbone part of the plasmid to be integrated into the genome. Furthermore, the recombination sites are separated by aproximately 2.2 kilobases, knocking out al least one gene. | ||
<br> | <br> | ||
| + | |||
| + | [[File: Construct Recombination sites position.jpg| 500px| center]] | ||
| + | |||
| + | <br> | ||
| + | |||
You can see their original construct design [https://2010.igem.org/Design_usu.html#Overview_of_Integration| here]. | You can see their original construct design [https://2010.igem.org/Design_usu.html#Overview_of_Integration| here]. | ||
| + | |||
<br> | <br> | ||
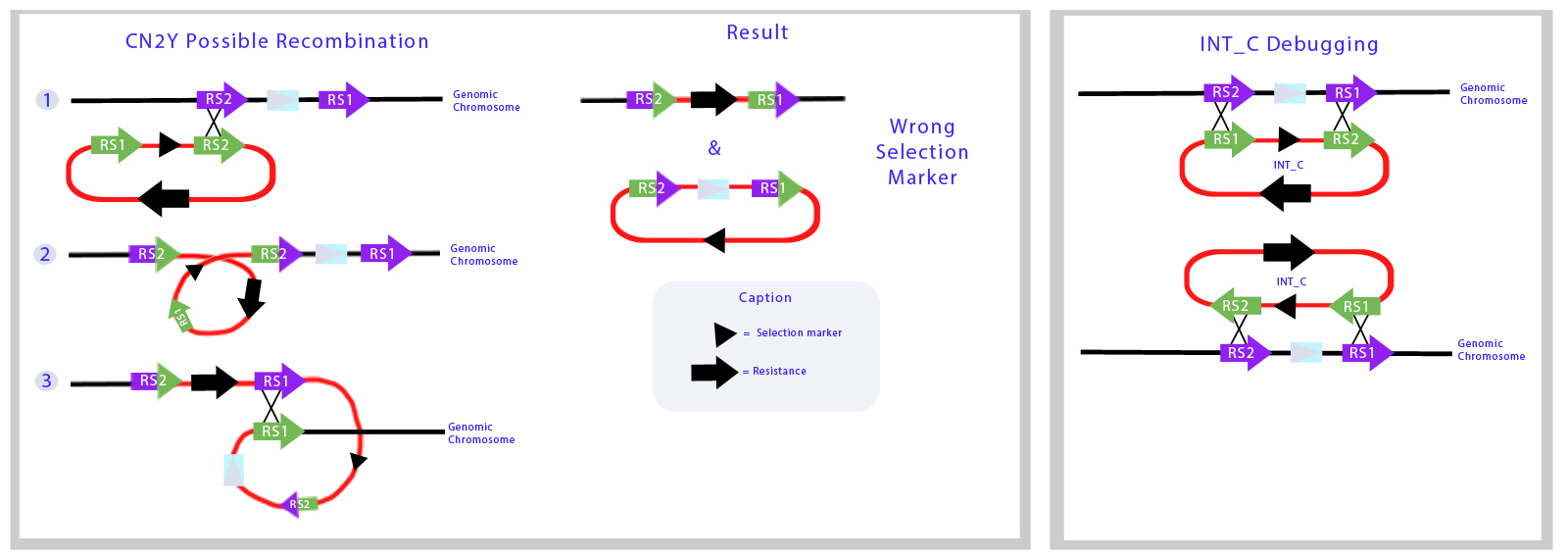
Here we illustrate the recombination mechanism in which the Int_C plasmid would integrate the backbone part of the vector to the genome. | Here we illustrate the recombination mechanism in which the Int_C plasmid would integrate the backbone part of the vector to the genome. | ||
| - | |||
| - | |||
[[File:UC_Chile-CN2Y_Possible_Recombination_Results_and_debugging.jpg|900 px|center]] | [[File:UC_Chile-CN2Y_Possible_Recombination_Results_and_debugging.jpg|900 px|center]] | ||
| + | |||
| + | <html> | ||
| + | <a href="https://2012.igem.org/Team:UC_Chile/Results/LuxBrick"><img src="https://static.igem.org/mediawiki/2012/a/ab/UC_Chile-Continue_button.jpg" align="right"> | ||
| + | </html> | ||
| + | |||
{{UC_Chilefooter}} | {{UC_Chilefooter}} | ||
Latest revision as of 01:18, 27 September 2012
 "
"