|
|
| (9 intermediate revisions not shown) |
| Line 2: |
Line 2: |
| | | | |
| | [[File: Chile_pipeting.jpg| 300px| right]] | | [[File: Chile_pipeting.jpg| 300px| right]] |
| - | <p>Here you will find various protocols we have used during the development of our project. We have separated them into 4 main sections: <br /><br /> [[#General Protocols]] <br /> [[#Cyanobacteria Protocols]] <br /> [[#Bactomithril Protocols]] <br /> [[#Characterization Protocols]] </p> | + | <p>Here you will find various protocols we have used during the development of our project. We have separated them into 3 main sections: <br /><br /> [[#General Protocols]] <br /> [[#Cyanobacteria Protocols]] <br /> [[#Characterization Protocols]] </p> |
| | <br /> | | <br /> |
| | <div id="General Protocols"> | | <div id="General Protocols"> |
| Line 268: |
Line 268: |
| | Concentrate the DNA to be transformed into Synechocystis to 1µg/µL in 10µL for each transformation (A total of 10 ug of the transformation plasmid). | | Concentrate the DNA to be transformed into Synechocystis to 1µg/µL in 10µL for each transformation (A total of 10 ug of the transformation plasmid). |
| | <br> | | <br> |
| - | Inoculate Synechocystis PCC 6803 into a sterile 250mL Erlenmeyer flask with 150mL* of liquid BG-11 and grow culture until you reach an OD730nm of 0.8 – 1.0 (That should take between 6 to 10 days dependending on the amount of initial inoculum). | + | Inoculate Synechocystis PCC 6803 into a sterile 250mL Erlenmeyer flask with 150mL* of liquid BG-11 and grow culture until you reach an OD730nm of 0.4 – 0.6, never above 0.8 (That should take between 6 to 10 days dependending on the amount of initial inoculum). |
| | * For each 50 mL of Synechocystis PCC 6803 you will have 5 individual transformations. | | * For each 50 mL of Synechocystis PCC 6803 you will have 5 individual transformations. |
| | <br> | | <br> |
| Line 317: |
Line 317: |
| | <div id="Reinoculation"> | | <div id="Reinoculation"> |
| | <h2>Reinoculation</h2> | | <h2>Reinoculation</h2> |
| - | Protocol right here!
| |
| - | </div>
| |
| | | | |
| - | <div id="Cyanobacterial DNA extraction"> | + | <br /> |
| - | <h2>Cyanobacterial DNA extraction</h2>
| + | This is a standard protocol for propagating cultures. The amount of initial innoculum will depend on when you will need a specific OD. Refer to [https://2012.igem.org/Team:UC_Chile/Results/Growthcurve growth curves] to calculate your needed inoculum through a standard C*V = constant relation. |
| - | </div>
| + | |
| - | <p>Protocol goes here!</p>
| + | |
| | | | |
| - | <div id="Bactomithril Protocol">
| |
| - | <h1>Bactomithril Protocols</h1>
| |
| - | </div>
| |
| | <br /> | | <br /> |
| - | <p>You can find a complete protocol to produce spider-silk fibers in the following paper:
| |
| | | | |
| - | Teulé, F., Cooper, A. R., Furin, W. a, Bittencourt, D., Rech, E. L., Brooks, A., & Lewis, R. V. (2009). A protocol for the production of recombinant spider silk-like proteins for artificial fiber spinning. Nature protocols, 4(3), 341-55. doi:10.1038/nprot.2008.250
| + | All work must be done under sterile conditions. |
| | | | |
| - | <br><br>Other easier protocols are in the following adresses:<br> | + | <br /> |
| | | | |
| - | http://www.pnas.org/content/107/32/14059.long<br>
| + | <ul> |
| - | | + | <li>Autoclave a flask with enough volume to harbor at least twice the volume you require</li> |
| - | http://onlinelibrary.wiley.com/doi/10.1002/jbm.a.34353/full</p>
| + | <li>Measure and add BG-11 media to flask</li> |
| | + | <li>Add axenic Synechocystis to flask. Inoculum must be in range 5-20% v/v of final volume</li> |
| | + | </ul> |
| | + | </div> |
| | | | |
| | <div id="Characterization Protocols"> | | <div id="Characterization Protocols"> |
| Line 345: |
Line 340: |
| | <div id="SyneGrowth"> | | <div id="SyneGrowth"> |
| | <h2>Methods for the characterization of Synechocystis PCC 6803 growth curve</h2> | | <h2>Methods for the characterization of Synechocystis PCC 6803 growth curve</h2> |
| - | This are the methods we used for setting up the growth curve experiment of our Synechocystis PCC 6803 which is further described [[Team:UC_Chile2/Characterization | here ]].
| + | These are the methods we used for setting up the growth curve experiment of our Synechocystis PCC 6803 which is further described [[Team:UC_Chile2/Characterization | here ]]. |
| | | | |
| | <h3>Materials</h3> | | <h3>Materials</h3> |
| Line 404: |
Line 399: |
| | <div id="Luciferase Assay"> | | <div id="Luciferase Assay"> |
| | <h2>Promoters Luciferase Assay</h2> | | <h2>Promoters Luciferase Assay</h2> |
| | + | We measured: |
| | + | (1) Luminiscence of LuxAB genes (from ''Xenorhabdus luminescens'' and ''Vibrio fischeri'') placed under [http://partsregistry.org/Part:BBa_K743003 Pta promoter] in Synechocystis upon induction with decanal and dodecanal. Both testing results at [http://partsregistry.org/Part:BBa_K743014:Experience] and [http://partsregistry.org/Part:BBa_K743015:Experience] |
| | + | (2) Luminiscence of [http://partsregistry.org/Part:BBa_K325905 Bacterial Lux reporter |
| | + | CDEG pλ AB] upon induction with decanal and dodecanal. Testing results [http://partsregistry.org/Part:BBa_K325905:Experience here] |
| | + | |
| | + | Controls used: |
| | + | |
| | + | Positive control: E. coli with arabinose-induced [http://partsregistry.org/Part:BBa_K325909 Luxbrick] |
| | + | </div> |
| | + | Negative control 1: E. coli wt cells |
| | + | </div> |
| | + | Negative control 2: Synechocystis PCC6803 wt cells |
| | + | |
| | + | Method |
| | + | |
| | + | E. coli and Synechocystis PCC 6803 transformed with the target constructs and negative controls were grown in liquid media at 37 °C (E. coli) and 30 °C (Synechocystis PCC 6803). |
| | + | |
| | + | Two hours after the beginning of the experiment, OD at 600 nm was measured for K325905 and Top10 in order to reinoculate at 0.24 OD (600 nm) in a total volume of 1.5 mL. |
| | + | |
| | + | Aftewards, LuxBrick was induced with Arabinose to a concentration of 3 mM and left in shaker at 30°C along with K325905 and Top10. |
| | + | |
| | + | After two hours, 100 µl of transformed bacteria and negative controls were inoculated in a 96 plate well. Every sample was inoculated in octuplicate. Luminiscence of the wells was measured by illuminometer. |
| | + | |
| | + | Several plate measurements were done to the plate with decanal and dodecanal at different concentrations (including 0 concentration). |
| | + | |
| | + | Note: This experience was done twice. Measurements were done at 14 and 16 hour of circadian rhythm in order to evaluate production of LuxAB under transaldolase promoter. |
| | + | |
| | + | Results at hour 14 |
| | + | |
| | | | |
| | <h1>References</h1> | | <h1>References</h1> |
| Line 413: |
Line 437: |
| | [2]Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO. (2009). "Enzymatic assembly of DNA molecules up to several hundred kilobases". Nature Methods 6 (5): 343–345. doi:10.1038/nmeth.1318 | | [2]Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO. (2009). "Enzymatic assembly of DNA molecules up to several hundred kilobases". Nature Methods 6 (5): 343–345. doi:10.1038/nmeth.1318 |
| | </div> | | </div> |
| | + | |
| | + | |
| | + | <html> |
| | + | <a href="https://2012.igem.org/Team:UC_Chile/Arduino"><img src="https://static.igem.org/mediawiki/2012/a/ab/UC_Chile-Continue_button.jpg" align="right"> |
| | + | </html> |
| | + | |
| | {{UC_Chilefooter}} | | {{UC_Chilefooter}} |

Here you will find various protocols we have used during the development of our project. We have separated them into 3 main sections:
#General Protocols
#Cyanobacteria Protocols
#Characterization Protocols
Growth media
LB media
Final concentrations:
- 1% (w/v) tryptone
- 0.5% (w/v) yeast extract
- 86 mM NaCl
Per liter:
- 10g Tryptone
- 5g Yeast extract
- 5g NaCl
- 15g Agar -Optional-
- Adjust pH to 7.5 with NaOH 1M
SOB media
Final concentrations:
- 0.5% (w/v) yeast extract
- 2% (w/v) tryptone
- 10 mM NaCl
- 2.5 mM KCl
- 20 mM MgSO4
Per liter:
- 5 g yeast extract
- 20 g tryptone
- 0.584 g NaCl
- 0.186 g KCl
- 2.4 g MgSO4
- 15g Agar -Optional-
- Adjust pH to 7.5 with NaOH 1M
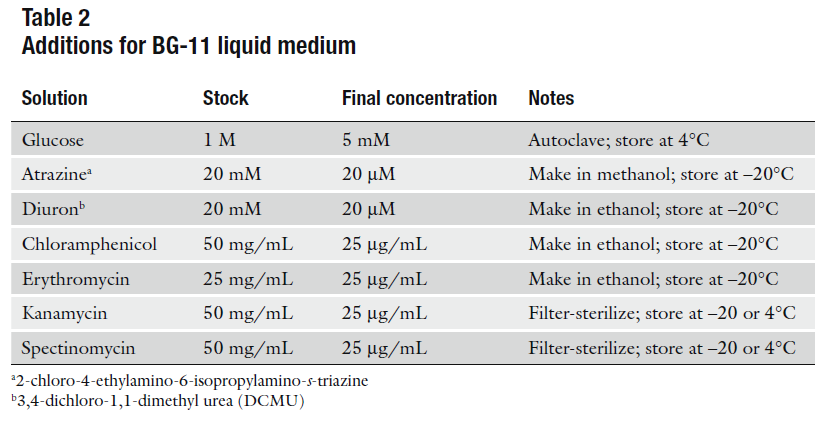
BG-11
For 1L:
- 1.5gr NaNO3
- 1L distilled H2O
- 10mL Concentrated salts solution 100X (4°C)
- 200µL H2PO4 1M (Room T°)
- 10gr BactoAgar -optional-
BG-110
For 1L:
- 1L distilled H2O
- 10mL Concentrated salts solution 100X (4°C)
- 200µL H2PO4 1M (Room T°)
- 10gr BactoAgar -optional-
BG-11C
For 1L:
- 1.5gr NaNO3
- 1L distilled H2O
- 10mL Concentrated salts solution 100X (4°C)
- 200µL H2PO4 1M (Room T°)
- 0.84gr NaHCO3
- 10gr BactoAgar -opcional-
Concentrated salts solution 100X
For 1L:
- 7.5gr SO4Mg * 7H2O
- 3.6gr CaCl2 * 2H2O
- 0.6gr Citric Acid
- 0.6gr Ammonium Ferric Citrate
- 0.093gr EDTA-Na2
- 2.0gr CO3 Na2
- 0.286gr BO3H3
- 0.181gr MnCl2 * 4H2O
- 0.022gr ZnSO4 * 7H2O
- 0.039gr NaMoO4 * 2H2O
- 0.0079gr CuSO4 * 5H2O
- 0.0049gr CoCl2 * 6H2O
- 1L distilled H2O
Autoclave and store at 4°C.
Buffers
5X Gibson Assembly Isothermal Buffer
Final Concentrations:
- 25% PEG MW 8000
- 500mM Tris HCl pH 7.5
- 50mM CaCl
- 50mM DTT
- 1mM dATP
- 1mM dTTP
- 1mM dGTP
- 1mM dCTP
- 5mM NAD+
- H20 nuclease free
For a 2 mL stock (equivalent to 20 100uL 5X isothermal buffers)
- 0.5g PEG MW 8000
- 1.5mL Tris-HCl pH 7.5 1M
Swivel for 30 minutes in a orbital oscillator for PEG to dissolve fully
- 50uL CaCl 2M
- 100uL DTT 1M
- 20uL dATP 100mM
- 20uL dTTP 100mM
- 20uL dGTP 100mM
- 20uL dCTP 100mM
- 200uL NAD+ 200mM
- H20 nuclease free up to 2mL
Alicuot into 100uL stocks
Store at -80°C
1.33X Gibson Assembly Master Mix
For a 375ul 1.33X Gibson Assembly Master Mix
- 100uL 5X Gibson Assembly Isothermal Buffer
- 6.25uL Phusion Polymerase 2 U/uL (cat N° F-350S) from Thermo Scientific
- 2uL T5 Exonuclease 1U/uL (cat N° T5E4111K) from Epicentre
It is important to dilute the T5 Exonuclease stock from 10U/uL to 1U/ul to measure the volume correctly
- 50uL Taq DNA Ligase 2000 U/uL (cat N° M208S) from NEB
- 216.75uL of nuclease free H20
Alicuot 9uL in 0.2mL PCR tubes. This will yield about 42 reactions
DNA assembly protocols
Standard Assembly
The standard assembly reaction relies on the standarization of Biobricks to join different DNA parts. Plasmids from the registry of standard parts allow joining of DNA parts by using a combination of specific enzymes to cut a Upstream Biobrick with the restriction enzymes EcoRI and SpeI, and a Downstream Biobrick with the restriction enzymes XbaI and PstI into a Destination plasmid which has been cut with the restriction enzymes EcoRI and PstI. The reaction yields the Upstream and Downstream part joined by a mixed restriction site and allows further elongation of the construct by the same strategy. It is important to notice that such a reaction requires purification of the digested parts if any of the plasmids (Upstream, Downstream or Destination) share a resistance marker.
- Upstream part enzymes: EcoRI & SpeI
- Downstream part enzymes: XbaI & PstI
- Destination backbone enzymes: EcoRI & PstI
Digestion reaction
- X volume of DNA to 500ng of plasmid
- (42.5 - X)uL of nuclease free water
- 5uL of NEB buffer 2
- 0.5uL of BSA 100X
- 1uL of Enzyme 1
- 1uL of Enzyme 2
- Incubate at 37°C for 2 hours
- Heat inactivate enzymes at 80°C for 20 minutes
Ligation reaction
- 2uL of digested Upstream part
- 2uL of digested Downstream part
- 2uL of digested Destination plasmid
- 2uL of T4 DNA ligase buffer
- 11uL of nuclease free water
- 1uL of T4 DNA ligase
- Incubate at room temperature for 10 minutes
- Heat inactivate at 80°C for 20 minutes
- Transform
Gibson Assembly
Gibson Assembly is a DNA assembly method created by Daniel Gibson during the development of the first Synthetic Genome (Synthia) 2. Its adaptation to a cloning method allows fast and accurate production of increasingly complex constructions. The strategy behind the method relies on PCR to obtain different parts which share a 40bp homology region, and a 3 enzyme reaction which produces cohesive ends, fills the gaps between the parts and ligates the resulting construct into a scarless assembly of various (>2) parts.
Primer design
The easiest way to design primers to obtain amplicons with the required overlaps (40bp final overlaps) is to make an in sillico design of the final construct
- Design forward primer of right amplicon of joint in 5'->3' direction
- Calculate length of annealing part of primer as to reach a Tm of approximately 63°C
- Add 20 bp of overlap
- Design reverse primer of left amplicon of joint in 5'->3' direction
- To do this, select full joint and "reverse complement" it (be sure to be able to discriminate between right and left parts of joint)
- Calculate length of annealing part of primer as to reach a Tm of approximately 63°C
- Add 20 bp of overlap
- Apply same principle in all joints
Obtaining parts
- PCR parts using Phusion Polymerase datasheet indications. It is important to use a low ammount of template plasmid (10pg) as to reduce possibility of carry-over during band purification
- Run agarose gel electrophoresis on a adecuate gel (50bp to 200bp parts should be run on a 3% w/v agarose gel, larger parts should be run on a 1% w/v to 1.5% w/v agarose gel) until clear distinction of bands is achieved. It is recommended that gels should be exposed as little as possible to UV transilluminators as UV light damages DNA.
- Cut band and proceed with band purification. Elute in smallest volume as possible according to your kit specification
- Quantify purified DNA
Assembly reaction
The assembly reaction is composed of 9uL of 1.33X Gibson assembly master mix + 3uL of purified parts DNA
- The calculation of the ratio at which your DNA parts are should be in correspondence to the level of competence of your cells. We have found that with our cells (5*10^⁸ colonies/ug of pUC19 DNA) the following ratios work well
- Calculate the amount of pmoles of each purified DNA part using the following equation: pmoles of DNA = weigth in ng * 1000 (conversion factor from nano to pico) / (650 Daltons * base pair length of part)
- We have seen that using 0.01 picomoles of template (part with the selection resistance) and the rest of parts at 0.03 picomoles and above, yield a high ratio of true positive transformants and a decent number of colonies to check
- Thoroughly mix the 3uL of purified parts DNA with the 9uL of 1.33X Gibson assembly master mix in ice
- Directly incubate the reaction at 50°C for 1 hour
- Transform competent cells with 8uL of assembled DNA (that leaves 4uL to check in an agarose gel if the reaction fails)
Checking assembly
- Check transformant colonies by colony PCR using primers for whole insert
- Grow positive colonies in media with corresponding antibiotic
- Miniprep colonies and digest plasmid with EcoRI & PstI restriction enzymes
- If checking protocols validate until now, proceed by sequencing to corroborate
E.coli Transformation
</div>
Chemically Competent cells
To prepare our competent cells we used a protocol we found in openwetware.org. Starting from commercial TOP10 cells as the starting inoculus for the whole protocol, we get competence levels close to 5*10⁸ cfu/ug pUC19 DNA
Openwetware protocol here
Cyanobacteria Protocols
Transformation of Synechocystis PCC. 6803
By iGEM - UC_Chile Team 2012 (Based on 1)
Materials
- Liquid BG-11 Media
- Solid BG-11 Media
- Sterile 250 mL Erlenmeyer flask
- Sterile 50 mL Falcon tube (2)
- Sterile 1.5ml Eppendorf tubes
- Petri Dishes
- Millipore Membrane Filters, 0.45um HA (HAWP09000)
- Pasteur pipette
- Antibiotics (according to your selection resistance)
- DNA for transformation
To be prepared previously
Concentrate the DNA to be transformed into Synechocystis to 1µg/µL in 10µL for each transformation (A total of 10 ug of the transformation plasmid).
Inoculate Synechocystis PCC 6803 into a sterile 250mL Erlenmeyer flask with 150mL* of liquid BG-11 and grow culture until you reach an OD730nm of 0.4 – 0.6, never above 0.8 (That should take between 6 to 10 days dependending on the amount of initial inoculum).
- For each 50 mL of Synechocystis PCC 6803 you will have 5 individual transformations.
Once you have reached the OD730nm and have enough DNA, proceed to transform.
Transformation
All subsequent actions are to be realized under sterile conditions.
Plate preparation
Prepare 50 mL BG-11 plates with no antibiotic for the transformation day.
Put Millipore membrane filters on BG-11 plates.
- 9.- After the 5 hours of recuperation, take the celular suspension and spread over the Millipore membrane. The suspension is to be spreaded carefully over the whole membrane using a sterile bended Pasteur pipete.
- 10.- Let the membranes dry before transfering the plates to the growth chamber.
- 11.- Transfer plates to the growth chamber putting the membrane facing up and leave at 30°C for 12 hours in light.
- 12.- Transfer membranes to BG-11 plates that contain half of the concentration of the antibiotic (12.5 ug/mL of antibiotic) and leave growing for 3 days at 30°C in light.
- 13.- After the 3 days have passed, transfer the membrane to BG-11 plates with full concentration of antibiotic (25ug/mL).
Colonies should appear between 1 to 2 weeks after the transformation. First you will see most of the cellular suspension dissapear and slowly small colonies should appear in the plate.
Reinoculation
This is a standard protocol for propagating cultures. The amount of initial innoculum will depend on when you will need a specific OD. Refer to growth curves to calculate your needed inoculum through a standard C*V = constant relation.
All work must be done under sterile conditions.
- Autoclave a flask with enough volume to harbor at least twice the volume you require
- Measure and add BG-11 media to flask
- Add axenic Synechocystis to flask. Inoculum must be in range 5-20% v/v of final volume
Characterization Protocols
Methods for the characterization of Synechocystis PCC 6803 growth curve
These are the methods we used for setting up the growth curve experiment of our Synechocystis PCC 6803 which is further described here .
Materials
- 4 sterile (autoclaved) 250 mL Erlenmeyer flasks
- 1 liter of BG-11
- 250 mL of BG-110
- Axenic culture of Synechocystis PCC 6803
- Sterile filtered 1 mL tips
- Spectrophotometer
- 5 1 mL Cuvettes
All procedures should be realized under sterile conditions, preferably in a laminar flow hood.
- Measure 150 mL of BG-11 with a sterile 50 mL Falcon tube and put into each of the 3 Erlenmeyer flasks.
- Measure 150 mL of BG-110 with a sterile 50 mL Falcon tube and put into the remaining Erlenmeyer flask.
- Inoculate 1 mL of axenic Synechocystis PCC 6803 with and OD730 of X to each flask (including the one with BG-110).
- Place flasks in rotating incubator and try to avoid leaving them out of the incubator for long when taking measurements
- Measure OD600 and OD730 twice a day (preferably with at least 8 hours of difference between measurements)
- Repeat measurements for 14 days
- Pick a single colony from a transformed plate of K325909. Grow in LB media with chloramphenicol 30ug/mL O/N at 30°C.
- Measure OD at 600nm, then dilute to OD600 = 0.6 with growth media following direct C*V ratio to obtain bacterial stock for experiments.
- Induce with 3mM (L)-Arabinose (final concentration)
- Alicuot 600uL of bacteria and add LB media with chloramphenicol 30ug/mL + additives and inducers up to 1.5mL. This will yield a 0.24 OD600nm.
- Grow at 30°C for 1.5 hours (or at chosen T° condition for characterization).
- Alicuot 100uL into 4 wells of a 96 well falcon plaque.
- Measure luminescence in a ThermoScientific Luminoskan Ascent Microplate Luminometer.
Methods for Gibson Assembly for small parts characterization
The experiment for the characterization of T5 exonuclease concentration in Gibson Assembly for small parts was set as follows:
- After the design of the segment to be assembled as described in the Characterization section, amplification of the parts was done using standard Phusion Polymerase PCR conditions but with a total amount of template of 0.05ng
- Bands where cut and purification of DNA from agarose gel was realized using a Promega's Wizard SV Gel and PCR Clean-Up System, final elution volume of 20 ul
- Purified DNA was quantified using a Nanodrop 2000
- According to concentration of DNA, the amount of pmoles/uL of each part was calculated using the following formulae:
pmoles of DNA = weigth in ng * 1000 (conversion factor from nano to pico) / (650 Daltons * base pair length of part)
- Volumes of each part were calculated to obtain a final amount of 0.0125 pmoles of the small part and 0.0025 pmoles of the backbone in a final volume of 3 uL (add nuclease-free water to reach 3 uL) for each reaction
- For the preparation of Gibson Assembly Master Mixes, different volumes of a 10X T5 exonuclease 1.33X Gibson Assembly Master Mix were mixed with T5 exonuclease-free 1.33X Gibson Assembly Master Mix until appropriate concentrations were reached. If mixing volumes of any of the Master Mixes was lower than 1 uL, then one of the diluted T5 exonuclease 1.33X Master Mixes was used for the mix.
- Gibson Assembly was done as described here, and transformation proceeded as described here
- Each of the concentration points of the experiment was done in triplicate
- Red colonies and white colonies were counted 20 hours post-transformation
</ul>
Promoters Luciferase Assay
We measured:
(1) Luminiscence of LuxAB genes (from Xenorhabdus luminescens and Vibrio fischeri) placed under Pta promoter in Synechocystis upon induction with decanal and dodecanal. Both testing results at [1] and [2]
(2) Luminiscence of [http://partsregistry.org/Part:BBa_K325905 Bacterial Lux reporter
CDEG pλ AB] upon induction with decanal and dodecanal. Testing results here
Controls used:
Positive control: E. coli with arabinose-induced Luxbrick
Negative control 1: E. coli wt cells
Negative control 2: Synechocystis PCC6803 wt cells
Method
E. coli and Synechocystis PCC 6803 transformed with the target constructs and negative controls were grown in liquid media at 37 °C (E. coli) and 30 °C (Synechocystis PCC 6803).
Two hours after the beginning of the experiment, OD at 600 nm was measured for K325905 and Top10 in order to reinoculate at 0.24 OD (600 nm) in a total volume of 1.5 mL.
Aftewards, LuxBrick was induced with Arabinose to a concentration of 3 mM and left in shaker at 30°C along with K325905 and Top10.
After two hours, 100 µl of transformed bacteria and negative controls were inoculated in a 96 plate well. Every sample was inoculated in octuplicate. Luminiscence of the wells was measured by illuminometer.
Several plate measurements were done to the plate with decanal and dodecanal at different concentrations (including 0 concentration).
Note: This experience was done twice. Measurements were done at 14 and 16 hour of circadian rhythm in order to evaluate production of LuxAB under transaldolase promoter.
Results at hour 14
References
[1]Julian J. Eaton-Rye , “Construction of Gene Interruptions and Gene Deletions in the Cyanobacterium Synechocystis sp. Strain PCC 6803 ”, Photosynthesis Research Protocols, Methods in Molecular Biology, vol. 684, DOI 10.1007/978-1-60761-925-3_22
[2]Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO. (2009). "Enzymatic assembly of DNA molecules up to several hundred kilobases". Nature Methods 6 (5): 343–345. doi:10.1038/nmeth.1318

 "
"