Team:UC Chile/Cyanolux/Project short
From 2012.igem.org
(Difference between revisions)
Juan Alamos (Talk | contribs) |
|||
| (36 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<h1>Main Goal:</h1> | <h1>Main Goal:</h1> | ||
| - | Our project consists on achieving bioluminescence controlled under circadian rhythms with long-term functionality. Our aim is to produce a bioluminescent cyanobacteria which lights up during dusk hours and | + | Our project consists on achieving bioluminescence controlled under circadian rhythms with long-term functionality. Our aim is to produce a bioluminescent cyanobacteria which lights up during dusk hours and regenerates the substrates during the day. |
<h2>Rationale:</h2> | <h2>Rationale:</h2> | ||
| - | The importance of Biological context in Synthetic Biology has been largely underestimated. We have addressed this issue by centering our project on enhancing functionality of a | + | The importance of Biological context in Synthetic Biology has been largely underestimated. We have addressed this issue by centering our project on enhancing functionality of a previously characterized Biobrick, LuxBrick, by placing it in a context which allows new features. |
| - | <html><center><img src="https://static.igem.org/mediawiki/2012/3/3f/Rationaleschemechile.jpg" align="middle" width=" | + | <html><center><img src="https://static.igem.org/mediawiki/2012/3/3f/Rationaleschemechile.jpg" align="middle" width="996"></center></html> |
<h3>Bioluminescence</h3> | <h3>Bioluminescence</h3> | ||
| Line 15: | Line 15: | ||
In 2010 the Cambridge iGEM team Biobricked the LuxBrick, a collection of genes from the Lux operon that incorporate both the Luciferase and the substrate production enzymes without regulation, allowing endogenous bioluminescence on E. coli. | In 2010 the Cambridge iGEM team Biobricked the LuxBrick, a collection of genes from the Lux operon that incorporate both the Luciferase and the substrate production enzymes without regulation, allowing endogenous bioluminescence on E. coli. | ||
| - | <html><center><img src="https://static.igem.org/mediawiki/2012/f/fc/Biolumilulilu.jpg" align=" | + | <html><center><img src="https://static.igem.org/mediawiki/2012/f/fc/Biolumilulilu.jpg" align="left" width="720"></center></html> |
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
<h3>Chassis</h3> | <h3>Chassis</h3> | ||
| Line 22: | Line 35: | ||
Coupling the endogenous circadian rhythms of this organism to the expression of the Lux genes will enable high-level functionality, through an automatically switching system that turns on bioluminescence only when needed. | Coupling the endogenous circadian rhythms of this organism to the expression of the Lux genes will enable high-level functionality, through an automatically switching system that turns on bioluminescence only when needed. | ||
| - | <html><center><img src="https://static.igem.org/mediawiki/2012/d/da/Synefeatures2.jpg" align=" | + | <html><center><img src="https://static.igem.org/mediawiki/2012/d/da/Synefeatures2.jpg" align="left" width="720"></center></html> |
| - | |||
| - | |||
| - | |||
| - | In turn, the production of these enzymes can be specifically set to any desired time of the day by fusing their CDSs to promoters controlled by the | + | |
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <h2>Strategy</h2> | ||
| + | The limiting step for the bacterial bioluminescent reaction is the substrate (n-decanal) concentration, therefore, to control light emission over time we decided to control it´s abundance in the cells, which in our model is a function of the substrates generation (by Lux C, D, E and G enzymes) and consumption (by the LuxAB luciferase). | ||
| + | <html><center><img src="https://static.igem.org/mediawiki/2012/f/fc/Dospromotorusi.jpg" align="middle" width="850"></center></html> | ||
| + | |||
| + | |||
| + | In turn, the production of these enzymes can be specifically set to any desired time of the day by fusing their CDSs to promoters controlled by the circadian rhythm. | ||
<h3>Mathematical Modelling</h3> | <h3>Mathematical Modelling</h3> | ||
Our model works as a “black box” in which the input takes the form of a specific hour of the day (i.e the hour on which you want your metabolite to reach maximal concentration) and the output is a couple of promoters from Synechocystis genome. | Our model works as a “black box” in which the input takes the form of a specific hour of the day (i.e the hour on which you want your metabolite to reach maximal concentration) and the output is a couple of promoters from Synechocystis genome. | ||
| - | It assumes that the | + | It assumes that the metabolite's production is controlled by enzymes under the control of promoter 1 and it´s degradation by enzymes under promoter 2. |
| - | + | For more details please check [https://2012.igem.org/Team:UC_Chile/Cyanolux/Modelling here] | |
| + | <html><center><img src="https://static.igem.org/mediawiki/2012/d/db/Blackbox.2.jpg" align="middle" width="660"></center></html> | ||
| + | |||
<h3>Wetlab strategy</h3> | <h3>Wetlab strategy</h3> | ||
| - | Having chosen the right promoters we set out to built our constructs to transform | + | Having chosen the right promoters we set out to built our constructs to transform Synechocystis. |
| - | As there weren´t straighforward tools to start working with in the registry (i.e characterized plasmids backbones, protocols, etc) we started from | + | As there weren´t straighforward tools to start working with in the registry (i.e characterized plasmids backbones, protocols, etc) we started from scratch. |
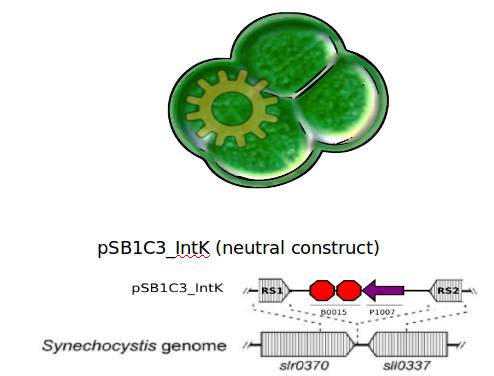
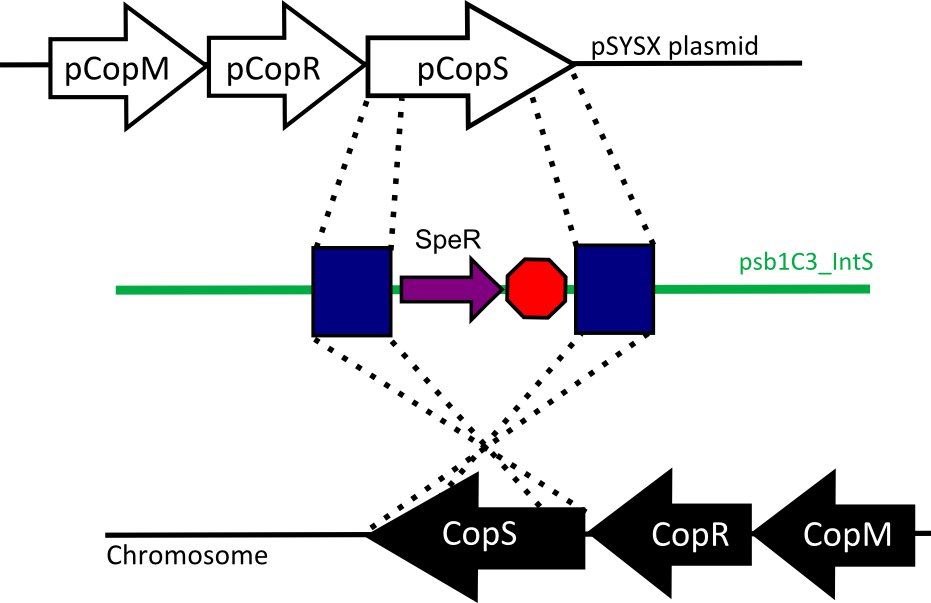
| - | We designed two recombination plasmids backbones | + | We designed two recombination plasmids backbones. One targets a gene essential for our chassis survival in the environment [https://2012.igem.org/Team:UC_Chile/Biosafety#Susceptibility_Construct (see biosafety)] and the other one a neutral site. |
| - | |||
| - | + | [[File:UC_Chile-IntKstrategy.jpg | 480px | left]] | |
| - | + | [[File:CopSmutants.jpg | 480px | right]] | |
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /><br /><br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| - | + | Our strategy was to insert the LuxAB genes (luciferase) under a circadian promoter with an expression peak right after dusk, in the neutral recombination plasmid; and the LuxCDEG genes (substrate production) with the appropriate promoter in the biosafety plasmid. | |
| - | + | See [https://2012.igem.org/Team:UC_Chile/Cyanolux/Results_short#Plasmid_construction plasmids in results section] | |
<h2>Implementation</h2> | <h2>Implementation</h2> | ||
| - | Synthetic biology inspires in nature | + | Synthetic biology inspires in nature making abstractions of its principles and mechanisms. We thought this moto could be applied beyond mollecular genetics... |
| - | We thought this moto could be applied beyond mollecular genetics. | + | |
With the relevance of context in mind, a biomimetic biolamp structure was designed that resembles the organ in which Vibrio fischeri -the bacteria from which the lux genes were biobricked- lives. | With the relevance of context in mind, a biomimetic biolamp structure was designed that resembles the organ in which Vibrio fischeri -the bacteria from which the lux genes were biobricked- lives. | ||
| + | <html><center><img src="https://static.igem.org/mediawiki/2012/b/b3/Biomimetic.jpg" align="middle" width="860"></center></html> | ||
| - | |||
| - | <div style="float: | + | [https://2012.igem.org/Team:UC_Chile/Cyanolux/Biolamp Full description of the biolamp device here] |
| - | [https://2012.igem.org/Team:UC_Chile/Cyanolux/Project See more about the project] | + | <div style="float:left"> |
| + | [https://2012.igem.org/Team:UC_Chile/Cyanolux/Project See more about the whole project] | ||
</div> | </div> | ||
| + | <br><br><br> | ||
| + | |||
| + | <html> | ||
| + | <a href="https://2012.igem.org/Team:UC_Chile/Cyanolux/Results_short"><img src="https://static.igem.org/mediawiki/2012/a/ab/UC_Chile-Continue_button.jpg" align="right"> | ||
| + | </html> | ||
| + | |||
{{UC_Chilefooter}} | {{UC_Chilefooter}} | ||
Latest revision as of 03:23, 27 October 2012
 "
"