Team:NTNU Trondheim/Logg
From 2012.igem.org
(→Thursday 28.05.12) |
|||
| (24 intermediate revisions not shown) | |||
| Line 194: | Line 194: | ||
</html> | </html> | ||
| + | ===Friday 06.07.12=== | ||
| + | Made a stock solution of 100 mM arabinose in H2O. The final concentration is slightly lower (by 10-15 %?) than 100 mM, as addition of 10 mL dH20 to the dry arabinose results in a final volume in excess of 10 mL. Sterilized the solution by filtering through a 0.2 um filter. Placed the stock solution in the refrigerator door shelf. | ||
| - | ===Thursday 28. | + | I suggest that we do a clean-up in the incubator and a review of all the cell cultures/agar plates that we have some day soon. --[[User:Jarlemag|Jarlemag]] 10:52, 6 July 2012 (CDT) |
| + | |||
| + | ===Thursday 05.07.12=== | ||
| + | Did a clean-up of the 37 C incubator cabinet. Discarded the following plates: | ||
| + | "Kan neg control 29.06" | ||
| + | "Neg ctrl 29.06" (chloramphenicol) | ||
| + | "LuxI 28.06" (Amp) | ||
| + | "LuxI Kloramfenikol 29.06" | ||
| + | "LuxI 29.06.12" (Kan) | ||
| + | "LI Amp BBa_K092400 28.06" | ||
| + | "Neg kontroll 28.06.12" (Amp) | ||
| + | "pBAD neg ctrl 29.6" (Amp) | ||
| + | |||
| + | Of the listed plates, only the following showed any growth: | ||
| + | |||
| + | "LuxI Kloramfenikol 29.06" (~15 colonies) | ||
| + | "LuxI 28.06" (Amp) (2 colonies) | ||
| + | "Neg ctrl 29.06" (chloramphenicol) (3 colonies) | ||
| + | |||
| + | It is reasonable to assume to attribute this late (and except for LuxI on Amp, in any case unexpected) growth to the break-down of the antibiotics after ~1 week at 37 C. --[[User:Jarlemag|Jarlemag]] 10:02, 5 July 2012 (CDT) | ||
| + | |||
| + | ===Wednesday 04.07.12=== | ||
| + | Performed restriction digest on the pBAD (<partinfo>BBa_K206000</partinfo>) and lysis <partinfo>BBa_K112808</partinfo> parts, cutting pBAD with SpeI and PstI, and the lysis part with XbaI and PstI. For pBAD, used 10 uL from isolated DNA sample "pBAD B1" (remeasured DNA concentration: 25 ng/uL). For the lysis part, used 2,3 uL from "lysis 1 DNA 24/6" (remeasured DNA concentration: 110 ng/uL). Incubated the restriction digest on waterbath at 37 C for aproximately 1.5 hour (protocol is 1h). No post-digestion CIP treatment or enzyme inactivation was performed. | ||
| + | |||
| + | Discarded the "Lysis 1" liquid culture inoculated on 1/7. Discarded 1 contaminated Erlen-Meyer bottle of LB. | ||
| + | |||
| + | ===Tuesday 03.07.12=== | ||
| + | Got started with our test constructs, and began by cutting an oxygenregulated promotor, VGB [http://partsregistry.org/Part:BBa_K561001], and RBS [http://partsregistry.org/Part:BBa_B0034]. We used the RBS-part as a backbone, and the VGB-promotor as the insert. | ||
| + | |||
| + | We tried running the VGB-promotor and RBS on gel, but the separation was poor, and we only got bands from the backbone. We then tried to cut the plasmids again, and added CIP to both of the solutions. | ||
| + | |||
| + | The solutions were then ligated [http://partsregistry.org/Help:Protocols/Ligation], transformed and plated out. | ||
| + | |||
| + | ===Monday 02.07.12=== | ||
| + | |||
| + | Restriction digestion was performed on isolated DNA from the VGB and RBS biobricks (<partinfo>BBa_k561001</partinfo> and <partinfo>BBa_B0034</partinfo>) according to [[Team:NTNU_Trondheim/Protocols|protocol]]. Gel electrophoresis was used to determine the size of the resulting fragments. The RBS restriction digest was filtered using the QIAquick PCR purification kit to remove the smallest fragment, which is expected to pass through the filter while the desired backbone is retained. The results for VGB were not as expected, and restriction digest of this part was therefore performed again, this time with addition of [http://en.wikipedia.org/wiki/Calf-intestinal_alkaline_phosphatase CIP] to the digest solution at the end of the procedure. | ||
| + | |||
| + | Both lysis 1 plates inoculated yesterday showed extensive growth (bacterial lawn). Moved them to the refrigerator. | ||
| + | |||
| + | ===Sunday 1.07.12=== | ||
| + | Miniprepped all three tubes with pBAD inoculated yesterday. Tube 3 contained 10 mL culture and was divided into two tubes, so that all processed tubes contained ~5 mL culture. Results of DNA measurements were as follows (ng/uL):<br> | ||
| + | pBAD B1: 24,0<br> | ||
| + | pBAD B2: 18,9<br> | ||
| + | pBAD B3-1: 11,4<br> | ||
| + | pBAD B3-2: 8,1 | ||
| + | |||
| + | Discarded B3-1 and B3-2, placed B1 and B2 in -20 C freezer. --[[User:Jarlemag|Jarlemag]] 11:04, 1 July 2012 (CDT) | ||
| + | |||
| + | In order to make a new plate of Lysis 1, transferred 20 uL from one of the tubes with liquid culture inoculated yesterday to one LA + Amp plate, and 50 uL to another. Placed the plates in the 37 C incubator cabinet. Also transferred 50 uL as inoculate to the tube used as negative control yesterday and placed it in the shaking incubator. Then combined all three tubes into one and placed in refrigerator to limit cell death before miniprep tomorrow. | ||
| + | --[[User:Jarlemag|Jarlemag]] 11:37, 1 July 2012 (CDT) | ||
| + | |||
| + | ===Saturday 30.06.12=== | ||
| + | |||
| + | Removed pBAD plates from 37 C incubator. Good growth on all transformed plates, Negative control showed no growth. Inoculated three colonies from plate B in liquid LB + Amp. Discarded the "mixed" plate. Placed plates A and B in refrigerator. Also inoculated three colonies from the lysis biobrick <partinfo>BBa_K112808</partinfo> using the plate from 22/6. Used an inoculating needle for all inoculations today. --[[User:Jarlemag|Jarlemag]] 11:03, 1 July 2012 (CDT) | ||
| + | |||
| + | ===Friday 29.06.12=== | ||
| + | |||
| + | Researched the ArcA protein and its binding sequence(s) to investigate whether the lldPRD operon promoter can be modified to eliminate repression by ArcA-P in the anaerobic state. Located a putative ArcA-P binding site in the promoter sequence: The sequence GTTAACTAAATGTTA is the reverse complement of the minus strand of the 85 bp promoter sequence from position 38 to 52. This sequence is identified by [http://onlinelibrary.wiley.com/doi/10.1046/j.1365-2958.1999.01347.x/full#t1 McGuire et al (1999)] (see [http://arep.med.harvard.edu/ecoli_matrices/dat/arcA.dat this] list). In the GenBank entry this part of the sequence reads TAACATTTAGTTAAC | ||
| + | |||
| + | Favorov et al. have made a newer suggestion for a general motif for ArcA binding sites - see [http://bioinformatics.oxfordjournals.org/content/21/10/2240.full#sec-5 this article] and the computational result [http://favorov.bioinfolab.net/SeSiMCMC/examples/arcA/result.html here]. According to Favorov, the crucial features of the site is a direct repeat, as shown below: | ||
| + | |||
| + | atacaTAACatttagtTAACcattc | ||
| + | |||
| + | Extracted the pBAD strong promoter biobrick <partinfo>BBa_K206000</partinfo> and transformed it in two samples. Plated out on three Amp plates as follows: 200 uL from sample A, 200 uL from sample B, 20 uL each from sample A and B. | ||
| + | |||
| + | --[[User:Jarlemag|Jarlemag]] 12:52, 29 June 2012 (CDT) | ||
| + | |||
| + | ===Thursday 28.06.12=== | ||
Researched the part <partinfo>BBa_K112805</partinfo> (subpart of Bba_K112808) containing the T4 Bacteriophage lysis t gene (locus T4p254 - [http://www.ncbi.nlm.nih.gov/nuccore/29366675?report=gbwithparts&from=160221&to=160877 GenBank entry]) encoding holin ([http://www.genome.jp/dbget-bin/www_bget?uniprot:VLYS_BPT4 Uniprot entry]) to determine if it must be assembled together with an RBS. The part sequence appears to have a RBS at bases 7-12 (AGGAG, which is the RBS [http://www.scielo.br/scielo.php?pid=S1415-47572004000400028&script=sci_arttext consensus sequence]) while the sequence found in GenBank (atg...) starts at base 17 of the part sequence. The recorded part sequence has a single base change, A->C, compared to the GenBank sequence, at position 70 (position 54 of the GenBank sequence). Otherwise, the part sequence is identical to the cited GenBank sequence, with the addition of the 16 preceding bases in the GenBank T4 complete genome sequence. For a review of the T4 genome, see http://mmbr.asm.org/content/67/1/86.long --[[User:Jarlemag|Jarlemag]] 05:34, 28 June 2012 (CDT) | Researched the part <partinfo>BBa_K112805</partinfo> (subpart of Bba_K112808) containing the T4 Bacteriophage lysis t gene (locus T4p254 - [http://www.ncbi.nlm.nih.gov/nuccore/29366675?report=gbwithparts&from=160221&to=160877 GenBank entry]) encoding holin ([http://www.genome.jp/dbget-bin/www_bget?uniprot:VLYS_BPT4 Uniprot entry]) to determine if it must be assembled together with an RBS. The part sequence appears to have a RBS at bases 7-12 (AGGAG, which is the RBS [http://www.scielo.br/scielo.php?pid=S1415-47572004000400028&script=sci_arttext consensus sequence]) while the sequence found in GenBank (atg...) starts at base 17 of the part sequence. The recorded part sequence has a single base change, A->C, compared to the GenBank sequence, at position 70 (position 54 of the GenBank sequence). Otherwise, the part sequence is identical to the cited GenBank sequence, with the addition of the 16 preceding bases in the GenBank T4 complete genome sequence. For a review of the T4 genome, see http://mmbr.asm.org/content/67/1/86.long --[[User:Jarlemag|Jarlemag]] 05:34, 28 June 2012 (CDT) | ||
| Line 204: | Line 273: | ||
''the lldPRD operon is proposed to be under the control of the global regulator ArcA, which binds to positions -14 to 3 with respect to the transcriptional start site, corresponding to the proposed P2 promoter, and represses the expression of this operon under anaerobic conditions.'' | ''the lldPRD operon is proposed to be under the control of the global regulator ArcA, which binds to positions -14 to 3 with respect to the transcriptional start site, corresponding to the proposed P2 promoter, and represses the expression of this operon under anaerobic conditions.'' | ||
| - | The cited reference is http://jb.asm.org/content/178/21/6238.full.pdf+html | + | The cited reference is http://jb.asm.org/content/178/21/6238.full.pdf+html (in this article the operon is called lctPRD) |
| + | |||
| + | <blockquote>''Of all the operons known to be regulated by the Arc system, | ||
| + | the lctPRD operon shows the widest range of response, with | ||
| + | 90- to 100-fold anaerobic repression apparent (23, 26'')</blockquote> | ||
| + | |||
| + | Of all the operons known to be regulated by the Arc system, | ||
| + | the lctPRD operon shows the widest range of response, with | ||
| + | 90- to 100-fold anaerobic repression apparent (23, 26) | ||
May this affect the usability of the promoter? --[[User:Jarlemag|Jarlemag]] 07:38, 28 June 2012 (CDT) | May this affect the usability of the promoter? --[[User:Jarlemag|Jarlemag]] 07:38, 28 June 2012 (CDT) | ||
If so, perhaps it could be possible to modify the sequence so that ArcA does not bind. | If so, perhaps it could be possible to modify the sequence so that ArcA does not bind. | ||
| - | ===Wednesday 27. | + | ===Wednesday 27.06.12=== |
Found the sequence for the lldp lactate operon at http://www.ncbi.nlm.nih.gov/nuccore/L13970 | Found the sequence for the lldp lactate operon at http://www.ncbi.nlm.nih.gov/nuccore/L13970 | ||
| Line 227: | Line 304: | ||
Some useful information on common laboratory strain mutations [http://books.google.no/books?id=OWXM4Q3-ieEC&pg=PA65&lpg=PA65&dq=lacZYA-argF&source=bl&ots=xqv8JrjVYZ&sig=P3MxMnbSDNh18yW-ek7kOGvmNec&hl=no&sa=X&ei=jPvqT5KsM-Tm4QSN3IT3Ag&ved=0CFsQ6AEwAg#v=onepage&q=lacZYA-argF&f=false here]. --[[User:Jarlemag|Jarlemag]] 08:11, 27 June 2012 (CDT) | Some useful information on common laboratory strain mutations [http://books.google.no/books?id=OWXM4Q3-ieEC&pg=PA65&lpg=PA65&dq=lacZYA-argF&source=bl&ots=xqv8JrjVYZ&sig=P3MxMnbSDNh18yW-ek7kOGvmNec&hl=no&sa=X&ei=jPvqT5KsM-Tm4QSN3IT3Ag&ved=0CFsQ6AEwAg#v=onepage&q=lacZYA-argF&f=false here]. --[[User:Jarlemag|Jarlemag]] 08:11, 27 June 2012 (CDT) | ||
| - | ===Tuesday 26. | + | ===Tuesday 26.06.12=== |
| Line 257: | Line 334: | ||
===Sunday 24.6=== | ===Sunday 24.6=== | ||
| - | Tube with culture inoculated from the 20 uL BBa_k112808 showed growth, despite the fact that no colonies were visible on the plate, and the "colony" that was scratched for inoculation turned out to be a water bubble below the agar. | + | Tube with culture inoculated from the 20 uL <partinfo>BBa_k112808</partinfo> showed growth, despite the fact that no colonies were visible on the plate, and the "colony" that was scratched for inoculation turned out to be a water bubble below the agar. |
| + | |||
| + | After miniprep of this culture, DNA concentration of two samples was measured with NanoDrop as 111 and 50 ng/uL. | ||
| - | + | Miniprepped culture of LuxR & HSL regulated promoter (<partinfo>BBa_R0062</partinfo>). Measured concentration of isolated DNA as 15,7 ng/uL. | |
| - | Extracted bricks <partinfo>BBa_B0034</partinfo> (abbreviated "RBS", Plate 1, well 2M) and BBa_B0015 (abbreviated "DTT", plate 1, well 23L) from the distribution kit for transformation. Stored the remainder of the resuspended BBa_B0034 DNA in the -20 C freeze. There was not enough DNA left over of BBa_B0015 to store. | + | Extracted bricks <partinfo>BBa_B0034</partinfo> (abbreviated "RBS", Plate 1, well 2M) and BBa_B0015 (abbreviated "DTT", plate 1, well 23L) from the distribution kit for transformation. Stored the remainder of the resuspended BBa_B0034 DNA in the -20 C freeze. There was not enough DNA left over of BBa_B0015 to store. --[[User:Jarlemag|Jarlemag]] 07:03, 5 July 2012 (CDT) |
===Saturday 23.06.12=== | ===Saturday 23.06.12=== | ||
Latest revision as of 15:52, 6 July 2012
Friday 06.07.12
Made a stock solution of 100 mM arabinose in H2O. The final concentration is slightly lower (by 10-15 %?) than 100 mM, as addition of 10 mL dH20 to the dry arabinose results in a final volume in excess of 10 mL. Sterilized the solution by filtering through a 0.2 um filter. Placed the stock solution in the refrigerator door shelf.
I suggest that we do a clean-up in the incubator and a review of all the cell cultures/agar plates that we have some day soon. --Jarlemag 10:52, 6 July 2012 (CDT)
Thursday 05.07.12
Did a clean-up of the 37 C incubator cabinet. Discarded the following plates: "Kan neg control 29.06" "Neg ctrl 29.06" (chloramphenicol) "LuxI 28.06" (Amp) "LuxI Kloramfenikol 29.06" "LuxI 29.06.12" (Kan) "LI Amp BBa_K092400 28.06" "Neg kontroll 28.06.12" (Amp) "pBAD neg ctrl 29.6" (Amp)
Of the listed plates, only the following showed any growth:
"LuxI Kloramfenikol 29.06" (~15 colonies) "LuxI 28.06" (Amp) (2 colonies) "Neg ctrl 29.06" (chloramphenicol) (3 colonies)
It is reasonable to assume to attribute this late (and except for LuxI on Amp, in any case unexpected) growth to the break-down of the antibiotics after ~1 week at 37 C. --Jarlemag 10:02, 5 July 2012 (CDT)
Wednesday 04.07.12
Performed restriction digest on the pBAD (<partinfo>BBa_K206000</partinfo>) and lysis <partinfo>BBa_K112808</partinfo> parts, cutting pBAD with SpeI and PstI, and the lysis part with XbaI and PstI. For pBAD, used 10 uL from isolated DNA sample "pBAD B1" (remeasured DNA concentration: 25 ng/uL). For the lysis part, used 2,3 uL from "lysis 1 DNA 24/6" (remeasured DNA concentration: 110 ng/uL). Incubated the restriction digest on waterbath at 37 C for aproximately 1.5 hour (protocol is 1h). No post-digestion CIP treatment or enzyme inactivation was performed.
Discarded the "Lysis 1" liquid culture inoculated on 1/7. Discarded 1 contaminated Erlen-Meyer bottle of LB.
Tuesday 03.07.12
Got started with our test constructs, and began by cutting an oxygenregulated promotor, VGB [http://partsregistry.org/Part:BBa_K561001], and RBS [http://partsregistry.org/Part:BBa_B0034]. We used the RBS-part as a backbone, and the VGB-promotor as the insert.
We tried running the VGB-promotor and RBS on gel, but the separation was poor, and we only got bands from the backbone. We then tried to cut the plasmids again, and added CIP to both of the solutions.
The solutions were then ligated [http://partsregistry.org/Help:Protocols/Ligation], transformed and plated out.
Monday 02.07.12
Restriction digestion was performed on isolated DNA from the VGB and RBS biobricks (<partinfo>BBa_k561001</partinfo> and <partinfo>BBa_B0034</partinfo>) according to protocol. Gel electrophoresis was used to determine the size of the resulting fragments. The RBS restriction digest was filtered using the QIAquick PCR purification kit to remove the smallest fragment, which is expected to pass through the filter while the desired backbone is retained. The results for VGB were not as expected, and restriction digest of this part was therefore performed again, this time with addition of [http://en.wikipedia.org/wiki/Calf-intestinal_alkaline_phosphatase CIP] to the digest solution at the end of the procedure.
Both lysis 1 plates inoculated yesterday showed extensive growth (bacterial lawn). Moved them to the refrigerator.
Sunday 1.07.12
Miniprepped all three tubes with pBAD inoculated yesterday. Tube 3 contained 10 mL culture and was divided into two tubes, so that all processed tubes contained ~5 mL culture. Results of DNA measurements were as follows (ng/uL):
pBAD B1: 24,0
pBAD B2: 18,9
pBAD B3-1: 11,4
pBAD B3-2: 8,1
Discarded B3-1 and B3-2, placed B1 and B2 in -20 C freezer. --Jarlemag 11:04, 1 July 2012 (CDT)
In order to make a new plate of Lysis 1, transferred 20 uL from one of the tubes with liquid culture inoculated yesterday to one LA + Amp plate, and 50 uL to another. Placed the plates in the 37 C incubator cabinet. Also transferred 50 uL as inoculate to the tube used as negative control yesterday and placed it in the shaking incubator. Then combined all three tubes into one and placed in refrigerator to limit cell death before miniprep tomorrow. --Jarlemag 11:37, 1 July 2012 (CDT)
Saturday 30.06.12
Removed pBAD plates from 37 C incubator. Good growth on all transformed plates, Negative control showed no growth. Inoculated three colonies from plate B in liquid LB + Amp. Discarded the "mixed" plate. Placed plates A and B in refrigerator. Also inoculated three colonies from the lysis biobrick <partinfo>BBa_K112808</partinfo> using the plate from 22/6. Used an inoculating needle for all inoculations today. --Jarlemag 11:03, 1 July 2012 (CDT)
Friday 29.06.12
Researched the ArcA protein and its binding sequence(s) to investigate whether the lldPRD operon promoter can be modified to eliminate repression by ArcA-P in the anaerobic state. Located a putative ArcA-P binding site in the promoter sequence: The sequence GTTAACTAAATGTTA is the reverse complement of the minus strand of the 85 bp promoter sequence from position 38 to 52. This sequence is identified by [http://onlinelibrary.wiley.com/doi/10.1046/j.1365-2958.1999.01347.x/full#t1 McGuire et al (1999)] (see [http://arep.med.harvard.edu/ecoli_matrices/dat/arcA.dat this] list). In the GenBank entry this part of the sequence reads TAACATTTAGTTAAC
Favorov et al. have made a newer suggestion for a general motif for ArcA binding sites - see [http://bioinformatics.oxfordjournals.org/content/21/10/2240.full#sec-5 this article] and the computational result [http://favorov.bioinfolab.net/SeSiMCMC/examples/arcA/result.html here]. According to Favorov, the crucial features of the site is a direct repeat, as shown below:
atacaTAACatttagtTAACcattc
Extracted the pBAD strong promoter biobrick <partinfo>BBa_K206000</partinfo> and transformed it in two samples. Plated out on three Amp plates as follows: 200 uL from sample A, 200 uL from sample B, 20 uL each from sample A and B.
--Jarlemag 12:52, 29 June 2012 (CDT)
Thursday 28.06.12
Researched the part <partinfo>BBa_K112805</partinfo> (subpart of Bba_K112808) containing the T4 Bacteriophage lysis t gene (locus T4p254 - [http://www.ncbi.nlm.nih.gov/nuccore/29366675?report=gbwithparts&from=160221&to=160877 GenBank entry]) encoding holin ([http://www.genome.jp/dbget-bin/www_bget?uniprot:VLYS_BPT4 Uniprot entry]) to determine if it must be assembled together with an RBS. The part sequence appears to have a RBS at bases 7-12 (AGGAG, which is the RBS [http://www.scielo.br/scielo.php?pid=S1415-47572004000400028&script=sci_arttext consensus sequence]) while the sequence found in GenBank (atg...) starts at base 17 of the part sequence. The recorded part sequence has a single base change, A->C, compared to the GenBank sequence, at position 70 (position 54 of the GenBank sequence). Otherwise, the part sequence is identical to the cited GenBank sequence, with the addition of the 16 preceding bases in the GenBank T4 complete genome sequence. For a review of the T4 genome, see http://mmbr.asm.org/content/67/1/86.long --Jarlemag 05:34, 28 June 2012 (CDT)
Found a reference indicating that the lldPRD operon is inhibited during anaerobic conditions (http://jb.asm.org/content/190/8/2997.full.pdf+html):
the lldPRD operon is proposed to be under the control of the global regulator ArcA, which binds to positions -14 to 3 with respect to the transcriptional start site, corresponding to the proposed P2 promoter, and represses the expression of this operon under anaerobic conditions.
The cited reference is http://jb.asm.org/content/178/21/6238.full.pdf+html (in this article the operon is called lctPRD)
Of all the operons known to be regulated by the Arc system, the lctPRD operon shows the widest range of response, with 90- to 100-fold anaerobic repression apparent (23, 26)
Of all the operons known to be regulated by the Arc system, the lctPRD operon shows the widest range of response, with 90- to 100-fold anaerobic repression apparent (23, 26)
May this affect the usability of the promoter? --Jarlemag 07:38, 28 June 2012 (CDT) If so, perhaps it could be possible to modify the sequence so that ArcA does not bind.
Wednesday 27.06.12
Found the sequence for the lldp lactate operon at http://www.ncbi.nlm.nih.gov/nuccore/L13970
Performed isolation of plasmid DNA from RBS (<partinfo>BBa_B0034</partinfo>, C78, C79 and <partinfo>BBa_K561001</partinfo> (VGF promoter). The yield from RBS was low today as well, so the spin column was centrifuged one more time together with the VGB-promoter.
Results from the nanodrop:
RBS: 30,2 ng/uL C78: 63,4 ng/uL C79: 70,5 ng/uL VGB: 22,9 ng/uL
Researched E. coli strains and their properties. Must DNA methylation be taken into account? Conclusion: No, not in the RFC 10 standard.
Some useful information on common laboratory strain mutations [http://books.google.no/books?id=OWXM4Q3-ieEC&pg=PA65&lpg=PA65&dq=lacZYA-argF&source=bl&ots=xqv8JrjVYZ&sig=P3MxMnbSDNh18yW-ek7kOGvmNec&hl=no&sa=X&ei=jPvqT5KsM-Tm4QSN3IT3Ag&ved=0CFsQ6AEwAg#v=onepage&q=lacZYA-argF&f=false here]. --Jarlemag 08:11, 27 June 2012 (CDT)
Tuesday 26.06.12
Colonies from C78, C79 and <partinfo>BBa_K561001</partinfo> (VGF promoter) were transferred from agar plates to liquid medium. There were two plates including the negative control that had not been incubated correctly (L1 and L2). They had not been placed with their lid facing downwards. These plates did not have any growth. The transformed cells from yesterday (L1 and L2) were then plated out onto new agar plates in hope to get colonies for further use.
Performed isolation of plasmid DNA from RBS (<partinfo>BBa_B0034</partinfo>, DTT (Short for "Double transcriptional terminator", <partinfo>BBa_B0015</partinfo>) grown on ampicillin and DTT grown on kanamycin. The yield was quite poor, especially for RBS. We then decided to run the spin columns one more time to see if we could get a higher yeild the second time. Since we are going to use a lot of RBS, we made a liquid culture right away, so we can perform a new DNA isolation on RBS tomorrow.
Results from the nanodrop:
RBS concentration: 16,8 ng/uL DTT + amp concentration: 35,8 ng/uL DTT + kan concentration: 26,5 ng/uL
Monday 25.06.12
Made chloramphenicol stock solution. Weighted out 0,135 g and dissolved in 4 mL 100 % ethanol to a concentration of 34 mg/mL. In media, a concentration of 24 ug/mL should be used. For this, 0,7 mL stock solution per liter medium is needed.
For todays transformations, used 0.1 mL supercompetent cells as there were no 0.5 mL tubes left.
Checked RBS and DTT plates from yesterday. No growth on negative control plate. 20++ on other three (RBS on La+Amp, DTT on La+Amp and La+ Kan).
For negative control when making liquid cultures, placed a clean toothpick in each of two tubes with LB + Amp and LA+ Kan respectively.
Placed the isolated DNA from the LuxR+HSL miniprep in the -20 C freezer.
Sunday 24.6
Tube with culture inoculated from the 20 uL <partinfo>BBa_k112808</partinfo> showed growth, despite the fact that no colonies were visible on the plate, and the "colony" that was scratched for inoculation turned out to be a water bubble below the agar.
After miniprep of this culture, DNA concentration of two samples was measured with NanoDrop as 111 and 50 ng/uL.
Miniprepped culture of LuxR & HSL regulated promoter (<partinfo>BBa_R0062</partinfo>). Measured concentration of isolated DNA as 15,7 ng/uL.
Extracted bricks <partinfo>BBa_B0034</partinfo> (abbreviated "RBS", Plate 1, well 2M) and BBa_B0015 (abbreviated "DTT", plate 1, well 23L) from the distribution kit for transformation. Stored the remainder of the resuspended BBa_B0034 DNA in the -20 C freeze. There was not enough DNA left over of BBa_B0015 to store. --Jarlemag 07:03, 5 July 2012 (CDT)
Saturday 23.06.12
Of two LA + Amp plates with <partinfo>BBaK112808</partinfo> transformants (20 and 200 uL) incubated since yesterday, only the 200 uL plate showed growth (10+ colonies). Negative control plate (untransformed cells) showed no growth. One colony was transferred to liquid culture.
On the 20 uL plate, a water bubble below the agar was mistaken for a colony, and a liquid culture was inoculated with a toothpick after scratching the plate. The mistake was realized, but the tube was still left to incubate, as it was thought it could act as a (weak) negative control.
Friday 22.06.12
The BBa_R0062 transformant from yesterday yielded several colonies on its plate, and one colony was inoculated into liquid medium and incubated at 37 C with shaking. After adding antibiotic to the liquid medium in the growth tube, some of the medium was spilled, so the growth volume was about 2 mL.
Isolation of plasmid DNA from all 7 initial transformants (see 20.06.12) was performed with the Promega Wizard Plus SV Minipreps DNA Purification System A1460.
The biobrick <partinfo>BBa_K112808</partinfo> (Enterobacteria phage T4 Lysis Device - no promoter) was extracted from the distribution kit and transformed into E. coli.
Thursday 21.06.12
We started the day with a lecture on genetic circuit modelling by Ph.d student and iGEM team instructor Marius Eidsaa, followed by a discussion of the options available in designing our system.
All 7 transformants from yesterday yielded 1 or more colonies. Negative controls (untransformed cells) plated out on Kanamycin and Ampicillin plates showed no growth, indicating that the antibiotics were effective in selecting transformants for growth. 1 colony from each of the transformants was inoculated in liquid LB medium and incubated at 37 C with shaking.
The biobrick [http://partsregistry.org/Part:BBa_R0062 BBa_R0062] (Promoter (luxR & HSL regulated -- lux pR)) was extracted from the distribution kit and transformed using our modified version of the official iGEM transformation protocol (see Protocols).
Wednesday 20.06.12
We started the day with an introduction to computer modeling of biological sytems, and use of the Cain chemical kinetics simulation program. We then researched and discussed various biobricks, and transformed several biobricks from the iGEM DNA distribution plates into E coli.
The following biobricks were extracted from the iGEM 2012 DNA distribution kit and transformed:
[http://partsregistry.org/partsdb/get_part.cgi?part=BBa_K091146 BBa_K091146]
[http://partsregistry.org/Part:BBa_K145150 BBa_K145150]
[http://partsregistry.org/partsdb/get_part.cgi?part=BBa_J23119 BBa_J23119]
[http://partsregistry.org/Part:BBa_E1010 BBa_E1010]
[http://partsregistry.org/Part:BBa_E0030 BBa_E0030]
[http://partsregistry.org/Part:BBa_E0020 BBa_E0020]
[http://partsregistry.org/Part:BBa_K082003 BBa_K082003]
For transformation, the iGEM HQ [http://partsregistry.org/Help:Protocols/Transformation transformation protocol] was followed, with the exception of the use of LB media in place of SOC and heat shock for 45 s instead of 60 s.
After media had been added to all tubes and the tubes placed on ice, it was discovered that the lid of the tube with Bba_E1010 was not closed tightly. The lid was then closed, and we believe that no media leaked out.
As this was the first time several of the team members performed the procedure, longer time than prescribed was spent on some parts of the protocol. This does not include the heat shock treatment, which is most time-critical.
Tuesday 19.06.12
We started the day preparing lab equipment. We sterilized pipette tips, toothpicks, water and LA and LB medium. We also prepared stock solutions of ampicillin and kanamycin, and made petri dishes containing LA + Amp (100 ug/mL) and LA + Kan (100 ug/mL). Equipment and solutions were autoclaved at 120 C for 20 minutes.
We prepared LB medium (1 L) and LA medium (2 L). The media were prepared by adding 1 L MQ water to the solid contents of each batch, giving a total volume slightly in excess of 1 L. We discussed whether we should instead add a smaller amount of water, dissolve the nutrients and then add water to an exact total volume of 1 L, but concluded that this would probably not be necessary.
After mixing, each batch was divided into two bottles for autoclaving. For the LA media, we had to retry this a couple of times, as the first times we were too slow: Some of the agar content solidified in the first bottle before pouring into the second, giving a lower agar content in the second bottle. We solved this problem by pouring all the liquid back into one bottle, reheating under stirring to redisolve the agar and then pouring the liquid quickly from one bottle to the other before the agar had time to solidify.
Stock solutions of Ampicillin and Kanamycin (both 100 mg/mL) were prepared from dry powders. After preparation and between use, the solutions were stored at -20 C.
We forgot to add a magnetic stirrer to one of the LA bottles before autoclaving, and thus hadd to add a magnet (flame sterilized) to the bottle afterwards, slightly increasing the chance of contamination.
After autoclaving, LA media bottles were left to cool. After reaching a temperature where they could be comfortably handled, the desired antibiotic was added using sterile technique. (500 uL stock solution to each bottle containing aproximiately 0.5 L, to a final antibiotic concentration of 100 ug/mL). The medium was then stirred and poured into petri dishes. After pouring, the petri dished were left to cool, and after solidifying enough for safe transport, placed in cold storage.
During addition of antibiotics to the media, two bottles of LA medium were confused, causing some doubt if Ampicillin had been added to one of the bottles or not. We believe that the problem was resolved correctly and that Ampicillin was added to both bottles. Still, if contamination becomes a problem with the ampicillin plates produced today, the question of whether all of them actually contain the antibiotic must be considered.
The above problem highlighted a challenge when working as a team in the lab and reinforced the importance of good labeling. We instituted a color coding system for antibiotics: All media and plates containing ampicillin should be marked with a green bar, while Kanmycin is indicated by a red bar.
Monday 18.06.12
The team visited the lab and was given a EHS run-through by Merethe Christensen. We also risk evaluated the project and handed in the risk assessment, so now we are ready to start working in the lab:-)
Wednesday 13.06.12
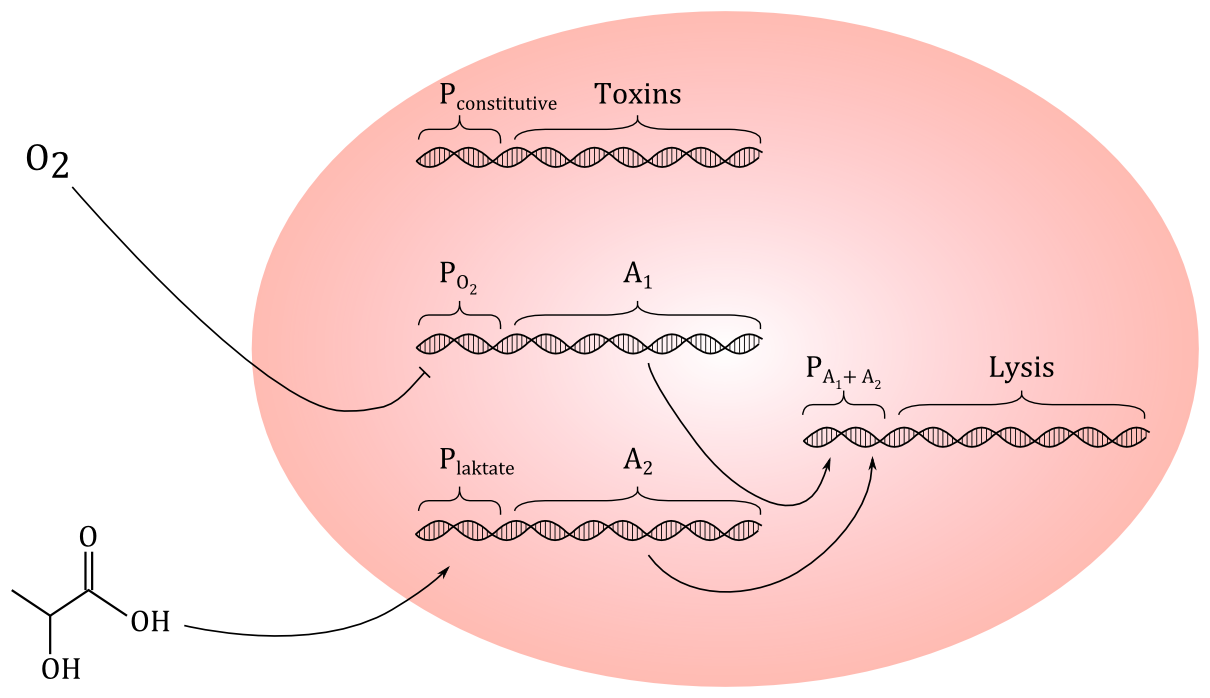
Today, we tried to come up with a preliminary genetic circuit. We decided that using HGF as a signal molecule could be difficult, since we don't even know if a protein this size could penetrate the outer membrane and the peptidoglycan layer of E.coli. But we have found out that cancer cells excrete more lactate than healthy cells, so we decided to go for lactate, which is a small molecule. If the construct works, it could be modified to respond to other signal molecules.
Preliminary genetic construct:
Friday 01.06.12
We talked about what we had found out since last week. Rolf and Jarle had since last week investigated different ways of making a cell lyse, and they found out that genes for lysis are already in this years iGEM kit. Ove and Nina had been investigating toxins, but didn't find as much as they hoped for, but they have however found some toxins. Colisin E1 should be possible to use. They have also sent an email to Pål Fallnes, who Marit Otterlei suggested we could try to get in touch with. Apparently, he has been working quite a lot with expression of toxins in bacteria. Gunvor and Eirin have since last week been trying to find a suitable signal molecule we could detect. Marit Otterlei suggested that we could use HGF, so Eirin and Gunvor did some reasearch around this growth factor. They found out that HGF is the only known lignad to the receptor c-Met, which is a tyrosin kinase. The idea so far is to use this, find out what signaling pathways this receptor is connected to, and find the endpoint of the signaling pathway. We assume that the endpoint is a growth factor possibly regulating a promoter, and if we find such a promoter, this could be set to control the lysis genes. The problem is that we don't know how signals are transducted to bacteria from the exterior, and this proved to be hard to find any information about. But we know that proteins resembling tyrosin kinases called BY kinases exists in bacteria, and also that proteins from eucaryotic cells have sometimes been working in bacteria when introns are removed by using mRNA and reverse transcriptase.
Both in the case of toxins we can express, and a signal molecule we could get our cells to respond to; we need more time. So we decided that Ove and Nina will continue looking for toxins for another week, and Eirin and Gunvor will investigate signal molecules more in depth.
Rahmi have found some possibly useful biobricks, and will be sending references to the biobricks in question to the team by email. Rahmi had also baked a cake for today's meeting:-)
In the case of sponsors, many of the companies we have asked request a budget. Gunvor will try to get hold of this from Eivind, and she will also talk to Merethe Christensen, who is an engineer at dept. of biotechnology, to arrange a safety excursion in the lab, which is necessary before we're allowed to work there.
We also elected Nina as our photo chief:-)
Friday 25.05.12
We started continuing our dicussion on which toxins we should make our cells produce. If we manage to make the cells lyse only in the presence of the signaling molecule we chose to go with, it shouldn't mather what the cells produce, but if the system turns out to be leaky, it will be a problem if the toxins we prodce is too toxic. That way we would also kill healthy cells. Also, another problem with too toxic molecules is that we will need a special lab to work with them. So we decided to go for the happy mean. We also discussed the posiibility of making the intracellular concentration of toxins a checkpoint for lysis, so that even if a signal molecule is present, or the cells is experiencing an oxygen deficient environment, they would still not lyse until the concentration of anti cancer molecules is high enough.
We now have three different modules involved in our project; a production part, a detection part and a lysis part. We decided to split in three groups and investigate these modules in depth;
- Production - Nina and Ove
- Nina and Ove will investigate further what kind of molecules we could make our cells produce, and since we got the names of some people working with toxins on the meeting with prof. Otterlei, we decided to contact them to hear if they can help us. We will also be contacting Jay Bradner.
- Detection - Eirin and Gunvor
- Eirin and Gunvor will look at possible signaling molecules we could use for detection of cancer cells. So far, we have decided to investigate HGF, the growth factor prof. Otterlei suggested, and VEGF.
- Lysis - Jarle and Rolf
- Jarle and Rolf will look at different ways to make a cell lyse, and they will also investigate the suicide switch used by the Tokyo team.
We also decided that we need a PR chief. Rolf volunteered!
Wednesday 23.05.12
We had our first meeting with our new advisor, Marit Otterlei, and we discussed which signal molecule we could use to detect the cancer cells in addition to using the O2 promoter to detect oxygen deficient areas. Prof. Otterlei suggested that we could use the Hepatocyte Growth Factor (abbreviated HGF), which is a growth factor regulating cell growth, cell motility and morphogenesis, that has the ability to bind to a tyrosine kinase. Prof. Otterlei also told us that many toxins can be produced by cells, so we decided to look more at making the cells produce toxins in addition to the ones we've already talked about.
Friday 18.05.12
We began this weeks meeting with going through the to-do-list from last meeting.
- Eirin:
- Has concacted Christopher Anderson by email to ask how his system works, but apparently he was unnwilling to give away too many details, as his work is not yet published.
- But we also came to the decision that specially because his work is still not published, we can't really be sure what his system looks like, and it is not our fault if it turns out that our systems actually work the same way, therefore, we shouldn't care too much that he is working on a similar project. But still; Rahmi and Eirin will try to get some more details out of him.
- Rolf and Jarle:
- Have been looking at possible sponsors. So far; we know that we will get a discount at VWR, so if we need something they sell, we should order it from them. Also, we have gotten a semi-positive response from Sigma-Aldrich.
- Before next meeting, Rolf will talk to more companies.
- Gunvor:
- Has since the last iGEM-meeting had a meeting with Gaute Brede, who is a postdoc working in the cell biology group at institute of cancer research and molecular medicine. I tried to ask him about signal molecules we could use, and his idea was to use ATP or adenosine, as cancer cells secrete more ATP than a normal cell. This is a signal molecule that attracts the attention of the immune system, so to avoid an imune reaction, the cancer cells also secrete enzymes called ectonucleotidases, which job is to degrade ATP to adenosine, which does not attract the immune system. Also healthy cells secrete ATP, but in smaller amounts than cancer cells. And healthy cells does not secrete CD39 and CD73, which is the ectonucleotidases responsible for degradation of ATP. So in summary; since cancer cells has these ectonucleotidases, they have lower levels of ATP in their surroundings compared to healthy cells, and higher levels of adenosine in their surroundings compared to healthy cells. Also, it seems like most types of cancer cells secrete CD39 and CD873.
We discussed the possibility of using adenosine as a signal molecule, but Rhami was sceptical, as he thinks adenosine and ATP are taken up by the cells through passive transport. So that means we might have to look for another signal molecule. Since we have few ideas ourself, we decided to have a meeting early next week instead, and try to get our new advisor Marit Ottelei to attend the meeting, since she might have some ideas.
In the time leading up to the next meeting, Rolf will continue looking for sponsors, and the ones who has time should start looking at different ways to lyse a cell.
Friday 11.05.12
We had our weekly meeting. We started by going through what we have done since last week:
- Eirin:
- Since last week, Eirin have been reading Christopher Andersons article. It seems like several parts of his project is similar to what we are planing now. But we still don't know if the cells Anderson used lysed by a stimulus given by the cancer cells. We have to find out if he did this or not, so before the next meeting, Eirin vil send an email to ask him.
- Ove:
- Ove will be looking more at making a calender function, so we can see a calendar on top of the page and be able to click different dates. Gunvor suggested that he could take a look at the code the NTNU team used to generate a calendar last year.
- Nina:
- Nina has found several new potential anti cancer drugs that can be syntheszed by cells, but she will be looking for more potential drugs in the time leading up to next meeting.
- Gunvor:
- Has asked around at Institute of Physics for cancer cells we could use in some of the experiments we are planning. She talked to the engineer responsible for the cell lab, and she said that they would like to provide us with cancer cells, but it would be an advantage if we could get some bottles of cancer cells from someone who is already using them. The problem is, she is going to leave for summer vacation in the middle of june, and if she's not here, we have to start our own cell line, which means that one of us will have to spend some hours a day in the cell lab attending to the cancer cells for about two weeks, which is the time it takes from the cells have been thawed to they are ready to be used in experiments.
- Since this solution doesn't seem optimal, Gunvor instead talked to a postdoc at her research group who has collaborations to researchers in the hospital. She has planned a meeting with one of her collaborators next wednesday, and Gunvor will follow her there to talk to them.
- She have also sent Marit Otterlei (professor at Institute of cancer research and molecular medicine) an email, and contacted Terje Espevik, who works with confocal microscopy at the hospital, about how we can characterize the O2-promoter using fluorescence microscopy.
We also decided that we should start looking for sponsors. We will submit an application for funds to Programme of Bioinformatics (PBI), and Rolf and Jarle is going to contact VWR, Sigma-Aldrich, and Fisher Scientific.
Eivind reminded us that it is important to come up with an idea for what our genetic construct will actually look like as soon as possible, so we can start the modelling. We decided that we will decide on an idea for a genetic circuit on next meeting, which will be on friday 18.05, at 13:30.
Have a nice weekend:-)
Wednesday 09.05.12
I have been playing around with a wiki design scheme today which can be found here. I hope we can discuss the wiki design a little bit this friday. Also I have been trying to make a calendar solution, but I haven't found any easier or more user-friendly way to implent this than to simply use the default wiki setup. So, at least for now, I think we should just keep using this site the way it is and add updates the way Gunvor did below (and I am doing now) ;) When you have added a new post to a day, you can click the button "Your signature with timestamp" in the editing menu to add your username along with the current time and date.
Hope you are all having a nice day and get to enjoy some sun!
--Oyas 09:45, 9 May 2012 (CDT)
Thursday 03.05.12
We had a meeting, and we discussed several things we would like to look more into before we start planning what our genetic circuit will actually look like.
Here is a list of what we decided to do, and who will do it:
- Check Christopher Anderson's article to see how similar his project was to ours - Eirin
- Make a calendar on our wiki page - Ove
- Look deeper into what kind of anti cancer drugs we could make our cells synthetize - Nina
- Check if we have access to cancer cells, and find out more about molecules that are secreted form cancer cells in larger amounts than by normal cells - Gunvor
- Find out more about the O2-sensitive promoter - Rahmi
- Rolf was also going to do something, but I have forgotten what he was going to do...
- We also decided to have our next meeting at 13:30 next friday, and we decided to eat lunch together on wednesdays
- Rahmi came with a suggestion for characterization of the O2-sensitive promoter:
- We could make the promoter control the expression of a fluorescent protein, then transform the system to cells, grow the cells on a soft medium they could migrate into, let them grow for a while, and then investigate the medium for coloured cells.
- Suggestion from Gunvor: We could use TIRF (Total Internal Reflection Fluorescence) microscope, which allows us to investigate the fluorescence in a certain layer of the medium.
Wednesday 02.05.12
Gunvor and Rahmi held an introductory lecture to cloning techniques. Gunvor held a crash course in molecular biology, the biobrick concept, and the most common molecular biology techniques, while Rahmi covered more advanced cloning techniques like SLIC and Gibson. If anyone wants more information on for example SLIC and Gibson, google j5 assembly;-)
Thursday 26.04.12
Today, all the groups from last time started with giving an overview of possible project within their topic. Then we discussed for quite a long time, and in the end we decided we wanted to work with cancer. But we also decided to keep the biosynthesis of fatty acids as a side project. The electricity project turned out to be quite hard to complete in only two months, so we decided to drop it.
Our final project idea is then to make bacteria, for example E.coli cells, produce anti cancer drugs; preferably as many different molecules as possible. We have read about both enzymes and endpoints of metabolic pathways that are disadvantageous for cancer cells. Our engineered cells should also be able to respond to a signal molecule secreted in larger amounts from cancer cells than from normal cells, for example a signaling molecule that promotes angiogenesis. When the cells detect the signal molecule from the cancer cells, they should lyse, releasing the anti cancer molecule close to the tumour. Another approach to reach the tumour could be to take advantage of the fact that the environment inside tumours normally is oxygen deficient. And as E.coli cells naturally migrate towards areas low in oxygen, a possible solution is also to activate the lysis gene when the cells are in an are with little oxygen. Rahmi told us about a promoter that gets activated by low concentration of O2. This could regulate the lysis genes.
For the main project, we have to make the cells lyse at a certain concentration of an outer stimuli. For our side project it could be interresting to do the oposite; making the cells lyse when they have produced a certain amount of fatty acids.
Friday 20.04.12
Today, we decided on the overall top three projects for the team. We added the points all the team members have given to the different projects, and the list then becomes as follows:
| Project | Number of points |
|---|---|
| Cancer (search and destroy) | 14 points |
| Biosynthesis of fatty acids | 10 points |
| Bacteria as electrical switch | 6 points |
| Water purification - hormones | 3 points |
| Water purification - sucralose | 1 point |
| Oil spill removing bacteria | 1 point |
| Biomining | 1 point |
We decided to look deeper into the overall top three projects. We formed three groups of two and two, and decided to look at the following things for the next meeting:
- Look at earlier projects on the topic
- Come up with some specific project ideas
- Look at the feasibility of the proposed projects
- Look at which biobricks we would need for each of the proposed projects, and see if they are present in the registry, or if we would have to make them ourselves.
The groups:
- Eirin and Jarle - Biosynthesis of fatty acids
- Rolf and Ove - Bacteria as electrical switch
- Nina and Gunvor - Cancer (search and destroy)
We also elected Ove as wiki chief:-)
Friday 13.04.12
Since we have quite a few project ideas to choose between now, we decided that for next meeting, all team members should pick their three favourite projects. We decided to give our favourite project three points, the project on second place two points, and our third favourite project one point. Before the next meeting we will add all the points together, and se which projects we should look at more in depth. At this meeting, we also discussed activities for Researchers Night. So far we have thought about making a construction kit to make the attendants on RN understand how we can combine different biobricks. We also discussed bringing pipettes, letting the attendants make alginate beads with different colours that they could bring home, and bringing a microscope and coloured cells (harmless, of course).
Friday 30.03.12
We kept discussing possible projects, and we now have several projects that would be interresting to work with. Here are the topics we have discussed so far:
- Fatty acid synthesis in E.coli (EDA and DHA production)
- Using cells in water purification
- Using bacteria to remove hormones from wastewater
- Using bacteria to remove sucralose from wastewater
- Be able to control pili production in order to control electron transfer between cells
- Biosensor detecting cancer cells, starting synthesis of chemoterapeutic drugs when cancer cells are detected
- Biosensor detecting harmful bacteria
- Making bacteria destroy oil spill
- Digitalize a signal from an environmental stimulus
- Biomining (using microorganisms to extract metals
Friday 16.03.12
We had a meeting, where we started discussing ideas for this year's iGEM project. We also decided to eat lunch together once a week, to get to know one another. For the outreach part of the project, Gunvor suggested that we could collaborate with Studentersamfundet to make a "lørdagsmøte" about synthetic biology.
Monday 27.02.12
We had our first meeting, and the team members met each other for the first time!
Friday 17.02.12
We recieved emails from Eivind letting us know that we are the ones that have been selected to represent NTNU in iGEM 2012. Everybody is happy!
 "
"