Team:Freiburg/Notebook/Toolkit
From 2012.igem.org
(→Labbook 1:) |
(→Labbook 1:) |
||
| Line 14: | Line 14: | ||
<p><br><br><br><br><br><br><br><br><br><br><br> | <p><br><br><br><br><br><br><br><br><br><br><br> | ||
Different colors indicate the six different primer pairs that were used during the PCR reaction | Different colors indicate the six different primer pairs that were used during the PCR reaction | ||
| - | The | + | The direpeats CC,CG and GC (yellow) were still missing from the synthesis order |
| - | We checked the | + | We checked the extension PCR with 4µl of 12 random direpeats and an additional template-missing control which was sitting in F1.</p><br> |
==<span style="color:#000000"> 9.7-15.7 == | ==<span style="color:#000000"> 9.7-15.7 == | ||
<p> | <p> | ||
| - | We digested AA and AC with Pst1 and Xba1 the digested direpeats were ligated in the psB1C3 | + | We digested AA and AC with Pst1 and Xba1; the digested direpeats were ligated in the psB1C3 vector and we tranformed 50 µl DH10B E.coli. The transformed E.coli were put on plates and incubated over night. |
| - | No colonies could be | + | No colonies could be detected and the experiment was redone without any growth</p><br> |
==<span style="color:#000000">16.7-22.9 == | ==<span style="color:#000000">16.7-22.9 == | ||
<p> | <p> | ||
| - | We figured out the problem of | + | We figured out the problem of last week: the primers we used for extensions were missing a piece at the end which made it impossible for the restriction enzymes to cut. We didn't want to order new primers, mainly because they are quite expensive, also it took the synthesis company three weeks last time and we were already late. As a new strategy we were trying to ligate our extensions into pJet blunt end vectors and start the restriction digest from this pJet plasmids. |
<br><br>We tried our new strategy with AA and AC and ligated them according to the pJet clonig vector protocol. We transformed DH10B bacteria with the ligated vectors and put them on plates. | <br><br>We tried our new strategy with AA and AC and ligated them according to the pJet clonig vector protocol. We transformed DH10B bacteria with the ligated vectors and put them on plates. | ||
| - | The grown colonies were picked for colony PCR and grown in liqud LB amp | + | The grown colonies were picked for colony PCR and grown in liqud LB amp medium. |
| - | We mini | + | We mini prepped the grown picks and used the prepped plasmids for digestion with Pst1 and Xba1</p><br> |
==<span style="color:#000000"> 23.7-29.7 == | ==<span style="color:#000000"> 23.7-29.7 == | ||
<p> | <p> | ||
| - | The digests were ligated | + | The digests were ligated in psB1C3 again and used for transforming DH10B. Once more we tried to plate them out but like the last time we didn't get any colonies. <br><br> |
| - | We | + | We reran the experiment and were able to get a few colonies. Analysis by colony PCR proved that the size of the insert was as expected. |
| - | We started to | + | We started to iterate the pJet strategy on the rest of our 96 direpeats to get them all to the point were they are inside the pJet vector and already stocked in DH10B</p><br> |
==<span style="color:#000000"> 30.7-5.8 == | ==<span style="color:#000000"> 30.7-5.8 == | ||
<p> | <p> | ||
| - | We spend the whole week ligating, | + | We spend the whole week ligating, transforming and picking colonies for colony pcr and liquid stocks. |
| - | <br><br> | + | <br><br>Also, we tried the digestion and ligation into the iGEM vector and got some colonies. The results of a colony PCR showed the right size. We started growing them in liqud LB and miniprepped them as our first finished Biobricks</p><br> |
==<span style="color:#000000"> 6.8- 12.8 == | ==<span style="color:#000000"> 6.8- 12.8 == | ||
<p> | <p> | ||
| - | After all the work | + | After all the work of last week, we got weird results from our colony PCR on some direpeats and chose to sequence some of our finished Biobricks. Sadly we found that what we thought were direpeats was nothing like the direpeats. After some sequence alining we could identify the problem as faulty design our extension primers. Because of the more or less matching sequences of the restriction sites, our extension primers formed not just dimers but trimers which created fragments with almost the exact same lenght of the expected direpeat. We checked some of our finished Biobricks and found a overwhelming number of them with just these primers inserted. With this information all of our finished Biobricks, as well as the finished direpeats in the pJet vector were thrown away. From colony PCR alone we could not be sure to have the right fragment. <br><br> |
As a conclusion we need to get a new strategy for creating our biobricks </p><br> | As a conclusion we need to get a new strategy for creating our biobricks </p><br> | ||
==<span style="color:#000000"> 13.8-19.8 == | ==<span style="color:#000000"> 13.8-19.8 == | ||
<p> | <p> | ||
| - | We had a | + | We had a lab meeting due to last week's problem. After discussing the problem we stopped the pJet strategy and came up with a somewhat weird approch. The problem we are facing is produced by the absence of basepairs directly after the outer restriction sites. To solve this we want to order a new pair of primers that fits on every end of the six different extension primers, which should not be a problem because they are all the same at 3' and 5'. With this new short primer pair we'll extend our already extended direpeats so that the restriction enzymes can now bind again and cut at the restrictions sites. This is much cheaper and also faster, because we can get such primers in one or two days instead of the 3 weeks for the long extension primers.<br><br> |
| - | We got the new Primers (so called ExEx Primers for extension of an extension) | + | We got the new Primers (so called ExEx Primers for extension of an extension) and started some testing to do a proof of concept experiment. After picking some clones we could see that our strategy worked out, but it didn't do as well as expected |
</p><br> | </p><br> | ||
==<span style="color:#000000">14.8-26.8 == | ==<span style="color:#000000">14.8-26.8 == | ||
<p> | <p> | ||
| - | We ran several | + | We ran several lines of experiments to get the perfect setup for our PCR and the ligation afterwards. After some setbacks, mostly with the ligation into the psb1c3 vector, we were able to create a complete protocol on how to get from direpeat to biobrick.<br><br> |
| - | After sequencing a first proof of concept | + | After sequencing a first proof of concept product from this new protocol, we now have our first six complete biobricks AG1-6.<br><br> |
| - | A new problem came up | + | A new problem came up: at the beginning of our project the synthesis products of our direpeats were cloned in pJet vectors and then stored in E.coli cryostocks. These stocks were grown in LB medium and miniprepped to get the direpeats. Because we had all these pJet issues, we sequenced all of the sixteen cryostocks and checked whether the direpats were really in there. Sadly, they were not. <br><br> |
| - | Good for us we still had some of the original synthesis product left and | + | Good for us, we still had some of the original synthesis product left and were able to start an amplification PCR.</p><br> |
==<span style="color:#000000"> 27.8-2.9 == | ==<span style="color:#000000"> 27.8-2.9 == | ||
<p> | <p> | ||
| - | The last month of the project started | + | The last month of the project started and we are now completely off our timetable. We started the toolbox from scratch and are at the lab now day and night. We are making some progress but slowly. The protocol we are using is designed so that there is no chance we are getting any false-positives. The digestion is done overnight and we are running all the digests on a gel to cut out the positive lanes. All of this takes time but we are now getting Biobricks.</p><br> |
==<span style="color:#000000"> 3.9-9.9 == | ==<span style="color:#000000"> 3.9-9.9 == | ||
<p> | <p> | ||
| - | First 35 biobricks are | + | First 35 biobricks are finished and sequenced, another 30 are in line waiting to be digested. We are a little short on hands in the lab but we are working to get it done. The homepage is also in the making, and we are positive to take at least the toolbox to the jamboree</p><br> |
==<span style="color:#000000">10.9-16.9 == | ==<span style="color:#000000">10.9-16.9 == | ||
<p> | <p> | ||
| - | Now we are at 70 biobricks and | + | Now we are at 70 biobricks and counting, the homepage is taking shape, we implemented dropdown navigation and a slidshow with pictures. With a little luck we can finish the toolbox this week.</p><br> |
==<span style="color:#000000"> 17.9-23. == | ==<span style="color:#000000"> 17.9-23. == | ||
<p> | <p> | ||
| - | No luck to report, the last direpeats CC and TA are messing with | + | No luck to report, the last direpeats CC and TA are messing with us. They just don't want to become Biobricks</p><br> |
==<span style="color:#000000"> 24.9-26.9 == | ==<span style="color:#000000"> 24.9-26.9 == | ||
<p> | <p> | ||
Revision as of 17:29, 24 September 2012
Labbook 1:
How we created the toolkit
2.7-8.7
Extension PCR of all the already synthesized direpeats on one plate
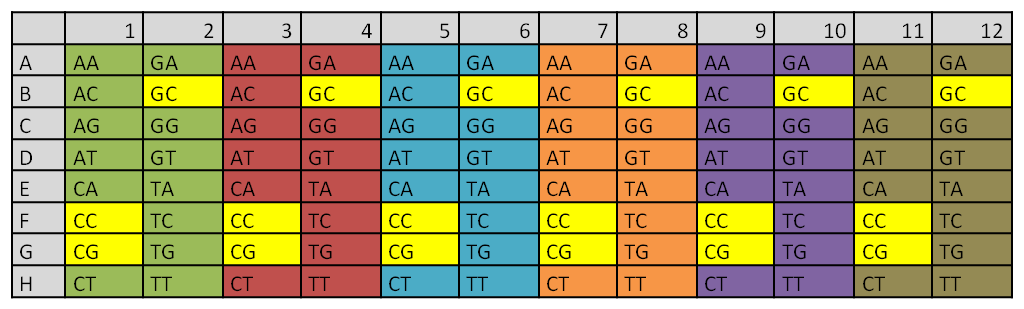
|
Different colors indicate the six different primer pairs that were used during the PCR reaction
The direpeats CC,CG and GC (yellow) were still missing from the synthesis order
We checked the extension PCR with 4µl of 12 random direpeats and an additional template-missing control which was sitting in F1.
9.7-15.7
We digested AA and AC with Pst1 and Xba1; the digested direpeats were ligated in the psB1C3 vector and we tranformed 50 µl DH10B E.coli. The transformed E.coli were put on plates and incubated over night. No colonies could be detected and the experiment was redone without any growth
16.7-22.9
We figured out the problem of last week: the primers we used for extensions were missing a piece at the end which made it impossible for the restriction enzymes to cut. We didn't want to order new primers, mainly because they are quite expensive, also it took the synthesis company three weeks last time and we were already late. As a new strategy we were trying to ligate our extensions into pJet blunt end vectors and start the restriction digest from this pJet plasmids.
We tried our new strategy with AA and AC and ligated them according to the pJet clonig vector protocol. We transformed DH10B bacteria with the ligated vectors and put them on plates.
The grown colonies were picked for colony PCR and grown in liqud LB amp medium.
We mini prepped the grown picks and used the prepped plasmids for digestion with Pst1 and Xba1
23.7-29.7
The digests were ligated in psB1C3 again and used for transforming DH10B. Once more we tried to plate them out but like the last time we didn't get any colonies.
We reran the experiment and were able to get a few colonies. Analysis by colony PCR proved that the size of the insert was as expected.
We started to iterate the pJet strategy on the rest of our 96 direpeats to get them all to the point were they are inside the pJet vector and already stocked in DH10B
30.7-5.8
We spend the whole week ligating, transforming and picking colonies for colony pcr and liquid stocks.
Also, we tried the digestion and ligation into the iGEM vector and got some colonies. The results of a colony PCR showed the right size. We started growing them in liqud LB and miniprepped them as our first finished Biobricks
6.8- 12.8
After all the work of last week, we got weird results from our colony PCR on some direpeats and chose to sequence some of our finished Biobricks. Sadly we found that what we thought were direpeats was nothing like the direpeats. After some sequence alining we could identify the problem as faulty design our extension primers. Because of the more or less matching sequences of the restriction sites, our extension primers formed not just dimers but trimers which created fragments with almost the exact same lenght of the expected direpeat. We checked some of our finished Biobricks and found a overwhelming number of them with just these primers inserted. With this information all of our finished Biobricks, as well as the finished direpeats in the pJet vector were thrown away. From colony PCR alone we could not be sure to have the right fragment.
As a conclusion we need to get a new strategy for creating our biobricks
13.8-19.8
We had a lab meeting due to last week's problem. After discussing the problem we stopped the pJet strategy and came up with a somewhat weird approch. The problem we are facing is produced by the absence of basepairs directly after the outer restriction sites. To solve this we want to order a new pair of primers that fits on every end of the six different extension primers, which should not be a problem because they are all the same at 3' and 5'. With this new short primer pair we'll extend our already extended direpeats so that the restriction enzymes can now bind again and cut at the restrictions sites. This is much cheaper and also faster, because we can get such primers in one or two days instead of the 3 weeks for the long extension primers.
We got the new Primers (so called ExEx Primers for extension of an extension) and started some testing to do a proof of concept experiment. After picking some clones we could see that our strategy worked out, but it didn't do as well as expected
14.8-26.8
We ran several lines of experiments to get the perfect setup for our PCR and the ligation afterwards. After some setbacks, mostly with the ligation into the psb1c3 vector, we were able to create a complete protocol on how to get from direpeat to biobrick.
After sequencing a first proof of concept product from this new protocol, we now have our first six complete biobricks AG1-6.
A new problem came up: at the beginning of our project the synthesis products of our direpeats were cloned in pJet vectors and then stored in E.coli cryostocks. These stocks were grown in LB medium and miniprepped to get the direpeats. Because we had all these pJet issues, we sequenced all of the sixteen cryostocks and checked whether the direpats were really in there. Sadly, they were not.
Good for us, we still had some of the original synthesis product left and were able to start an amplification PCR.
27.8-2.9
The last month of the project started and we are now completely off our timetable. We started the toolbox from scratch and are at the lab now day and night. We are making some progress but slowly. The protocol we are using is designed so that there is no chance we are getting any false-positives. The digestion is done overnight and we are running all the digests on a gel to cut out the positive lanes. All of this takes time but we are now getting Biobricks.
3.9-9.9
First 35 biobricks are finished and sequenced, another 30 are in line waiting to be digested. We are a little short on hands in the lab but we are working to get it done. The homepage is also in the making, and we are positive to take at least the toolbox to the jamboree
10.9-16.9
Now we are at 70 biobricks and counting, the homepage is taking shape, we implemented dropdown navigation and a slidshow with pictures. With a little luck we can finish the toolbox this week.
17.9-23.
No luck to report, the last direpeats CC and TA are messing with us. They just don't want to become Biobricks
24.9-26.9
 "
"
