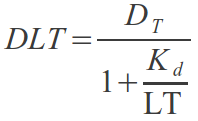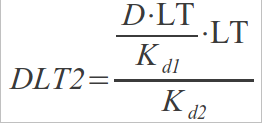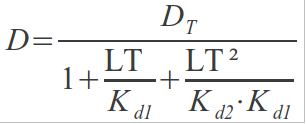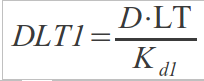Contents |
Why?
From our work on LOV2 photoactivation we should be able to predict the percentage of LovTAP-VP16 proteins that are photoactivated depending on the irradiance applied.
- Can we then predict the occupancy of TrpO binding sites in our reporter plasmid by LovTAP-VP16 dimers?
- And the difference in expression levels between an irradiated cell culture a one in the dark?
What?
Well, things start to get complicated, since the number of unknowns is huge in this problem, because it's affected by the transient transfection process, the cell's metabolism, the interaction of plasmids with the genomic DNA, the interaction of VP16 with the transcriptional cofactors and lots more.
We can split this into two parts:
- Estimation of the proportion of active plasmids that will be bound to LovTAP-VP16. We will assume the same binding affinity ratio (light/dark) as Strickland et al (2008) found for LovTAP at 25ºC because, although the absolute affinity is temperature dependent, according to Jin et al (1993), this change is not important. We are not sure whether only one or two LovTAP-VP16 dimer can bind a TrpO sequence. According to Yang et al (1996) a whole TrpO sequence can be bound by a tandem of two TrpR dimers. We will modelize both cases.
- Estimation of the expression level of reporter gene, dsRed in this case, depending on the proportion of bound reporter plasmids, based on the estimation from the previous part. Warning! This might give us an idea of the orders of magnitude, but can't be validated until lots of experiments have been done.
How?
To modelise the behavior of LovTAP-VP16, we will start with the following assuptions:
- [LovTAP-VP16] >> [Plasmids] in the nucleus.
- LovTAP-VP16 is always a dimer and the number of activated LOV2 domains is equivalent to the number of active dimers.
In our model, plasmids can be either free or bound to LovTAP-VP16, with different transcription rates for each state. The resulting network can be seen in diagram 1:
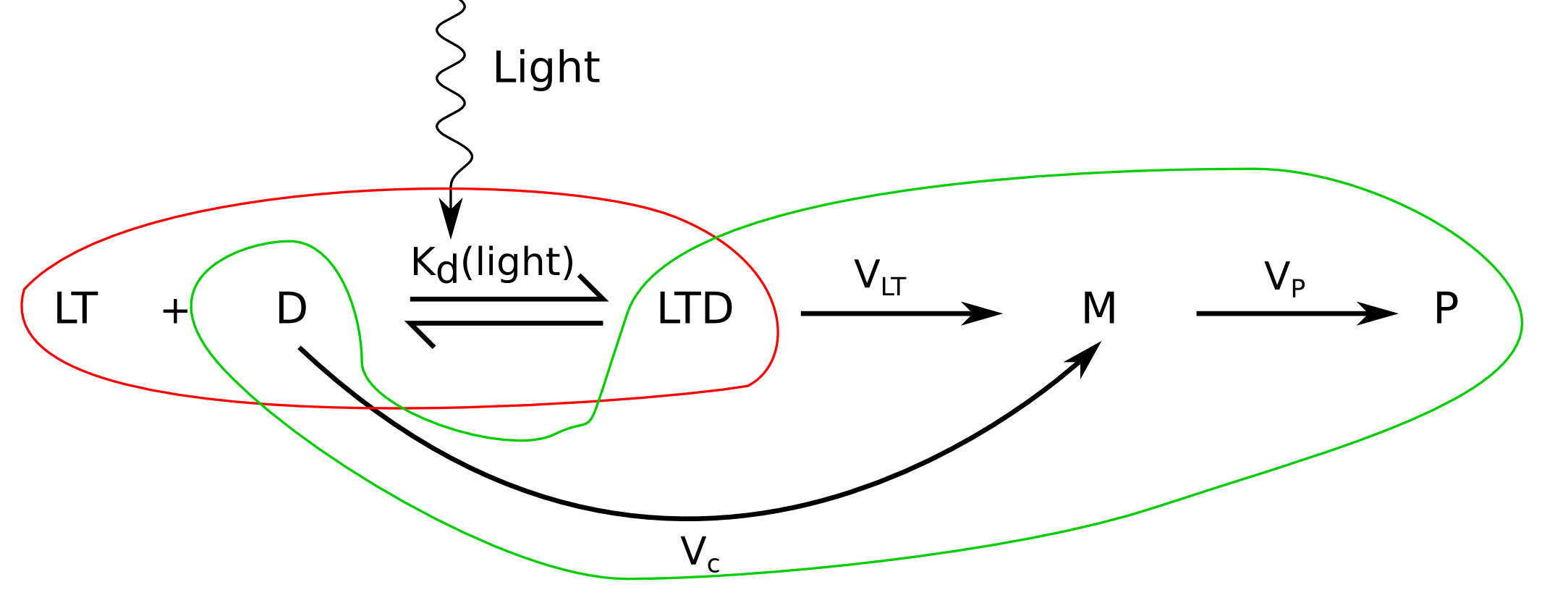
Photoactivation level
We will then assume that we know the proportion of activated LovTAP-VP16 monomers, since we have calculated it in this section. But LovTAP-VP16 has to dimerize to be able to bind DNA. To illustrate the importance of this, let's give an example: imagine a solution with LovTAP-VP16 where 50% of the monomers are activated. Assuming that all of them form part of a dimer, we get that 25% of the dimers are inactive, 25% active and 50% half active. The average binding affinity will mostly depend on the affinity of the half active half inactive dimers. In the section on the 3D protein structure of LovTAP-VP16 we conclude that it is reasonable to think that a half activated dimer will have a DNA activity somewhere between a fully activated dimer and a deactivated one. This means that we can take the monomer activation level to approximate the dimer activation level. Eitoku et al (2005) say it takes 2 ms for the Jα-helix to undock. This is 3 orders of magnitude faster than the photoactivation rate we expect with our setup. Therefore, we can consider the undocking instantaneous and thus say that photoactivation equals active LovTAP-VP16. Nonetheless, we might have to add an efficiency factor, since Yao et al (2008) found that 92% of photoactive LOV2 has it's Jα-helix undocked.
Number of activated reporter plasmids
Supposing a single binding site
Let's go step by step; if we assume that only one LovTAP-VP16 dimer can bind the TrpO binding site, reporter plasmids can exist in two states; free or bound to a LovTAP-VP16 dimer, and the total number of available plasmids must be the sum of the two:
According to the law of mass-action, the proportion of each one will also depend on the number of LovTAP-VP16 dimers floating in the nucleus. This proportion is called K_d, the dissociation constant, defined as:
with [D] the concentration of free binding sites, equal to the number of read-out plasmids not bound by LovTAP-VP16, [LT] the concentration of free LovTAP-VP16 dimers and [DLT] the concentration of plasmids with LovTAP-VP16 bound to them.
Since the volume we are interested in is a CHO cell's nucleus, we can suppose it approximately constant and get rid of the concentration units:
If we reorder, we get
what can be substituted in the first expression to get
Supposing a double binding site
In E. coli, TrpR can bind the TrpO sequence as a tandem of 2 dimers (Yang et al, 1996) What if there is place for 2 LovTAP-VP16 dimers in our TrpO binding site?
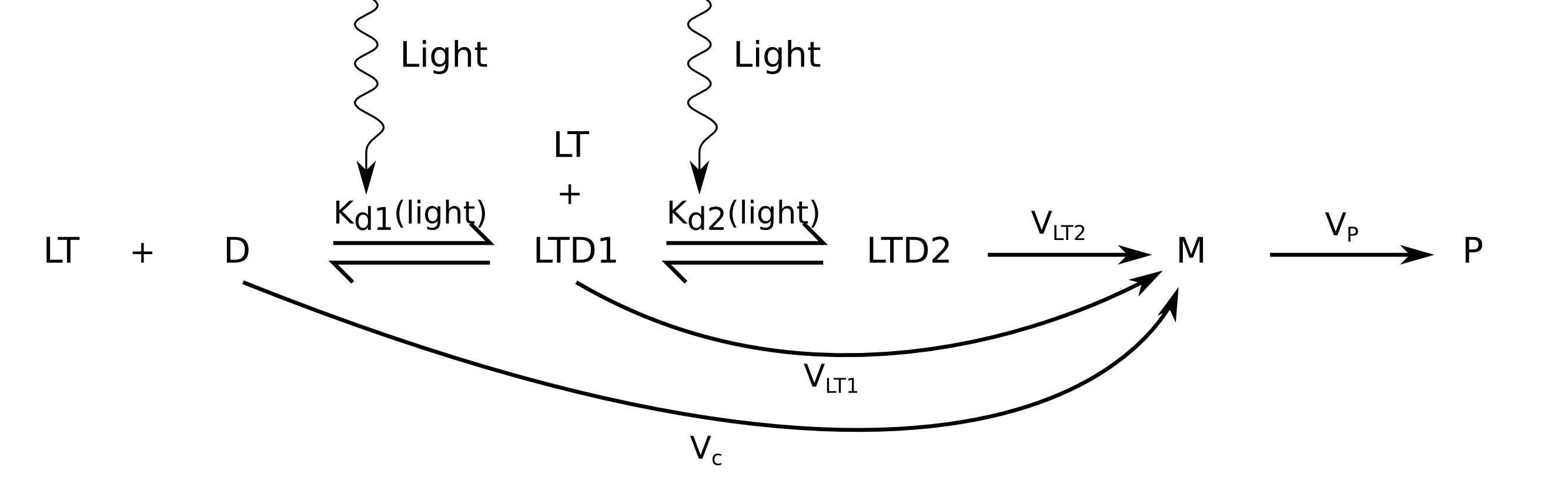
In this case, a binding site can be in 3 different states: free, bound to one dimer or bound to two dimers. Now the following expression should hold:
We have now two dissociation constants:
Performing the same operations as before:
Combining them:
Substituting DLT1 into DLT2:
And substituting DLT1 and DLT2 in the first expression:
Some steps of reordering:
If we substitute this in the following:
We get:
And finally:
Parameters and conclusions
If we just want to know the light/dark ratio of binding site occupancy, the concentration of reporter plasmids has no effect, so one parameter less. The Kd(light) = 142 ± 61 and Kd(dark) = 788 ± 94 nM have been set by Strickland et al (2008) for nano molar concentrations, up to 690 nM. Both Kd seem to depend on the concentration, with a maximum Kd(dark)/Kd(light) around 400 nM. This means that in fig. 1 only the areas of the curves around the maximum are reliable and the rest has a higher value than it would in reality. As we see, the curve which maximum is nearer to 400 nM is the one assuming double binding site and 30% activity of LTD1, or the half activated LovTAP-VP16 dimer. This is in agreement with the conclusion we got after visually inspecting the 3D structure of LovTAP-VP16.
Expression levels
Single binding site
We now know, more or less, the number of reporter gene copies that will be activated by LovTAP-VP16 and those that will be expressing only at constitutive levels.
The number of reporter mRNA copies in the cell will vary according to both transcription rates and the mRNA degradation rate:
And the number of reporter proteins in the cell will change depending on the number of mRNA copies and the protein degradation rate:
Double binding site
We have the same sort of expression as before, but we might have two different transcription activation rates with one or with two LovTAP-VP16 dimers binding a site:
Parameters
So far, we have been transfecting our CHO cells transiently using [http://en.wikipedia.org/wiki/Polyethylenimine PEI]. Following our protocol, we add 1.5 µg of DNA per million cells; that is 1.5x10⁻⁶ µg per cell. Our plasmids are about 6000 bp, or around 10⁻¹¹ µg per plasmid. This means we dose some 1.5x10⁵ plasmids per cell. How many can we expect, in average, to reach the nucleus? Well, experience in the lab shows that only some 15% to 30% of the cells show expression. According to Cohen et al, 2009, we could expect somewhere between 100 and 1000 plasmids to reach the nucleus in average. We don't know how many of those will be transcribed, and we don't know how the conditions in the nucleus will affect the way LovTAP-VP16 binds DNA.
From now on we can't expect producing more than qualitative information.
 "
"