Team:Chalmers-Gothenburg/Methods
From 2012.igem.org
m |
|||
| Line 1: | Line 1: | ||
{{Team:Chalmers-Gothenburg/Template2}} | {{Team:Chalmers-Gothenburg/Template2}} | ||
<div align="justify" style="width:auto; margin-left:30px; margin-right:30px;"> | <div align="justify" style="width:auto; margin-left:30px; margin-right:30px;"> | ||
| - | + | ||
| - | + | '''Notation!''' This page is under construction --[[User:Annaru|Annaru]] 10:09, 3 July 2012 (CDT) | |
| + | =Methods= | ||
| + | |||
| + | The work required for the construction of the biosensor is divided into three main tasks: (1) expression of the LH/CG receptor, (2) knock-out of the ''CWP2'' gene encoding a cell wall mannoprotein and (3) expression of indigo synthesizing enzymes tryptophanase and monooxygenase. These tasks will be performed in parallel and in the end, the different parts will be combined into one system. The three main tasks are described in detail in the following sections. | ||
| + | |||
| + | ==Expression of the LHCGR gene in yeast strain IMFD-73== | ||
| + | The first task is to express the LH/CG receptor in yeast. The receptor contains several amino acid residues that are essential for efficient ligand binding, both in the extracellular and in the transmembrane domains. In addition, the C-terminus and the intracellular loop III are considered to bind to the associated Gα protein of the receptor, and thereby constitute an important part of the signal transduction [[#Ryu|[1]]]. In order to ensure correct conformational change following ligand binding and to guarantee a proper signal transduction, the full-length LH/CG receptor will be expressed. The LHCGR gene will be inserted into the yeast strain IMFD-73 and be placed under the control of a strong, constitutive promoter to ensure constant expression. To enhance translation efficiency, a Kozak sequence (AAAACA) will also be inserted upstream of the LHCGR gene. In addition, a signal peptide will be added to the N-terminus of the receptor sequence to ensure transportation to the yeast cell membrane. | ||
| + | |||
| + | Previous research does not provide general guidelines for which signal peptide should be used when expressing human GPCRs in yeast. In this project, both the native signal of the LH/CG receptor, consisting of its 22 first amino acids [[#Dufau|[2]]], and a yeast signal peptide will be tested separately. The 5' end of the LH/CG receptor, which will be used as the native signal peptide, is originally glycosylated and glycosylation in yeast differs significantly from the one in human [[#Hamilton|[3]]]. The native signal will, however, be tested even though it is not sure if glycosylation of these amino acid residues is required for correct transportation of the receptor to the membrane [[#Petaja|[4]]]. In order to test a yeast signal, the idea was to fuse the signal peptide of Ste2, the receptor to be replaced, with the LH/CG receptor N-terminus. However, Ste2 lacks an identified signal sequence [[#Data|[5]]]. Instead, the signal peptide from Ste3, the MATα type pheromone receptor, will be used. It consists of the first 25 amino acid residues of the receptor [[#Data|[5]]]. | ||
| + | |||
| + | ==Plasmid construction== | ||
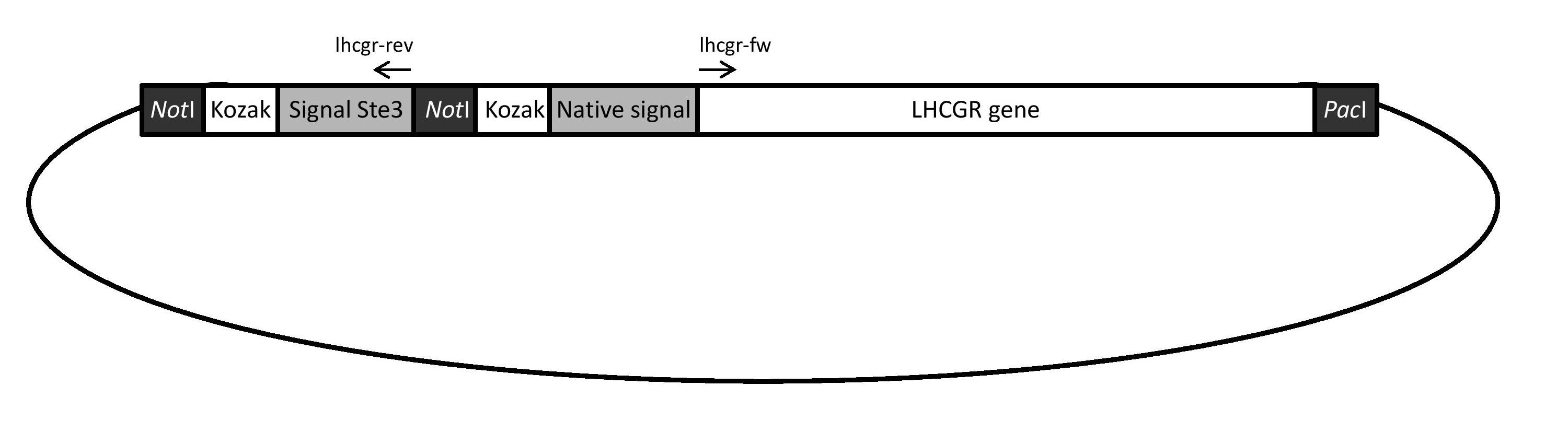
| + | The LHCGR gene construct will be ordered in a vector. Figure 1 provides a schematic illustration of this construct. The signal peptide of Ste3 and a Kozak sequence will be fused with a Kozak sequence at the 5' site of the native signal and the gene. In addition, ''Not''I and one ''Pac''I restriction sites will be added as indicated in Figure 1. | ||
| + | |||
| + | [[File:Genbild.jpg|center|700px|]] | ||
| + | <p align="justify"; style="width:auto; margin-left:80px; margin-right:80px;"><font face="verdana"><small>''' Figure 1: Schematic illustration of the LHCGR gene construct. ''' Using this construct enables expression of the LHCGR gene with each signal peptides separately. For the expression of the gene with the native signal, the vector can be digested directly with ''Not''I and ''Pac''I. When testing the expression with the yeast signal, PCR can be run with 5' phosphorylated primers indicated by arrows in order to exclude the native signal and its associated Kozak sequence and PacI restriction site. The resulting linear fragment can be religated. </small></font></p> | ||
| + | |||
| + | The aim of using this particular construct is to enable expression of the LHCGR gene with both signal peptides separately. For the expression with the native signal, the vector containing the LHCGR gene will be digested directly with ''Not''I and ''Pac''I restriction enzymes. The fragments will be purified on a gel and then ligated into the corresponding sites of the expression plasmid pSP-GM1 with bidirectional promoters TEF1 and PGK1, a ''URA3'' marker gene and an ampicillin resistance gene. For expression of the LHCGR with the Ste3 signal peptide, PCR will be run with 5' phosphorylated primers indicated with arrows in Figure 1 (lhcgr-rev and lhcgr-fw). This will result in a linear fragment that lacks the native signal and its associated ''Not''I site and Kozak sequence. This fragment will be purified on a gel, religated and amplified in ''E. coli''. After that, it will be digested with ''Not''I and ''Pac''I restriction enzymes, column purified and then ligated into the expression plasmid pSP-GM1. | ||
| + | |||
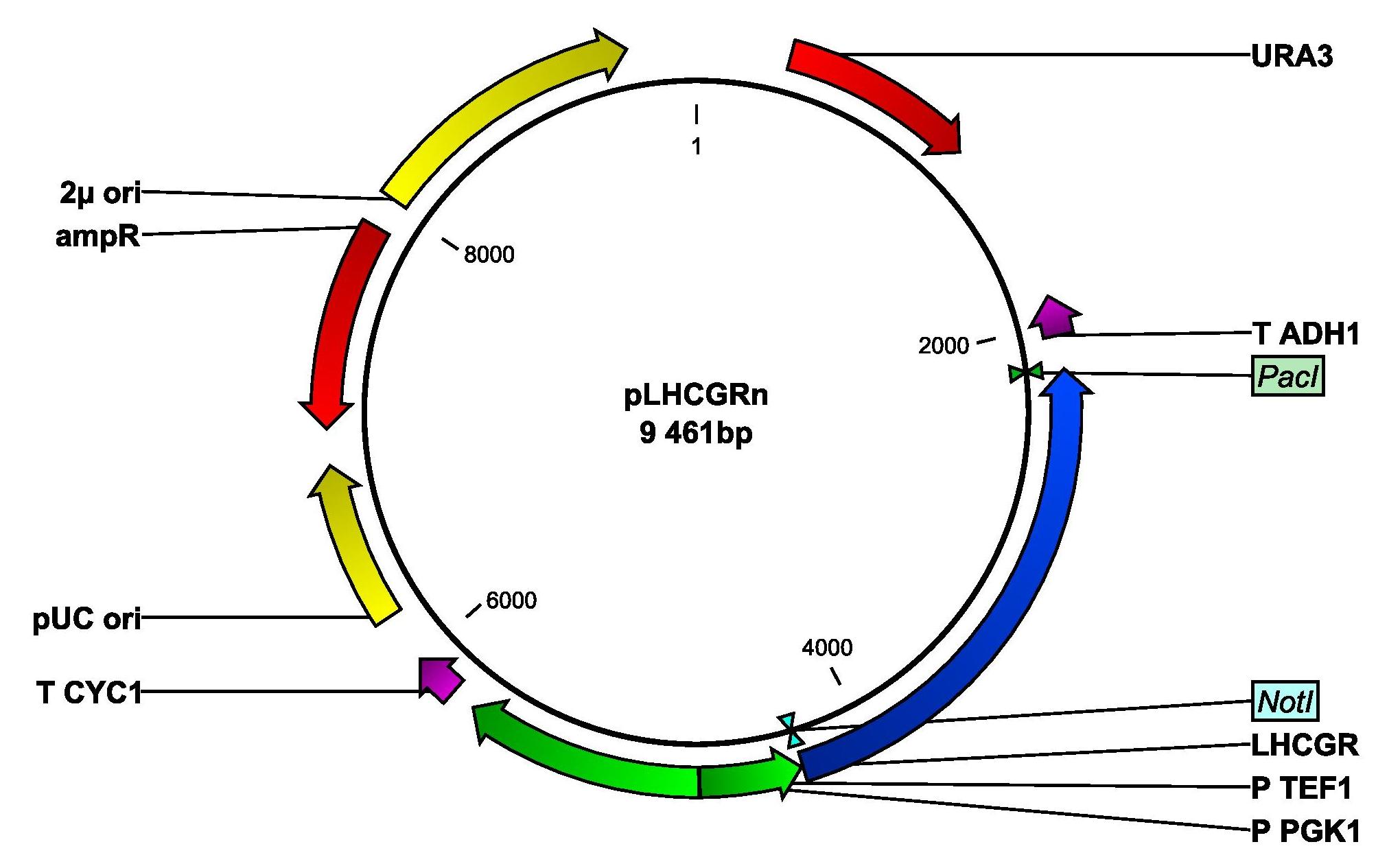
| + | In both cases, the gene will be set under the control of the strong and constitutive TEF1 promoter. The resulting plasmids will be named pLHCGRn (native signal peptide) and pLHCGRy (yeast signal peptide). Figure 2 shows a map of the plasmid pLHCGRn. pLHCGRy is similar to pLHCGRn and only differs in length of the LHCGR gene construct, which is 9 bp longer in the pLHCGRy plasmid. For amplification, ''E. coli'' strain DH5α will be transformed separately with the two plasmids and an empty control plasmid by heatshocking. The cells will be cultivated in ampicillin-containing LB medium. | ||
| + | |||
| + | [[File:HLHCGR.jpg|center|400px|]] | ||
| + | <p align="justify"; style="width:auto; margin-left:80px; margin-right:80px;"><font face="verdana"><small>''' Figure 2: Plasmid map of pLHCGRn derived from pSP-GM1. ''' The plasmid contains a yeast origin of replication, 2μ ori, and a yeast marker, ''URA3''. The parts that are important for transforming ''E. coli'' are the ampicillin resistance gene, AmpR and the E. coli origin of replication, pUC ori. The LHCGR gene construct with the yeast signal peptide is anked by ''Not''I and ''Pac''I. P TEF1 is a strong, constitutive promoter and T ADH1 is a terminator. </small></font></p> | ||
| + | |||
| + | |||
| + | =References= | ||
| + | <div id="Ryu"></div>[1] Ryu KS, Ji L, Chang L, Ji TH. Molecular mechanism of LH/CG-receptor activation. Molecular and Cellular Endocrinology. 1996;125(1-2):93-100. | ||
| + | |||
| + | <div id="Dufau"></div>[2] Dufau ML. The hormone luteininizing hormone receptor. Annual review of Physiology. 1998;60(1):461-496. | ||
| + | |||
| + | <div id="Hamilton"></div>[3] Hamilton SR, Gerngross TU. Glycosylation engineering in yeast: the advent of fully humanized yeast. Current Opinion in Biotechnology. 2007;18(5):387-392. | ||
| + | |||
| + | <div id="Petaja"></div>[4] Petaja-Repo UE, Merz WE, Rajaniemi HJ. Significance of the carbohydrate moiety of the rat ovarian luteinizing-hormone/chorionic-gonadotropin receptor for ligand-binding specificity and signal transduction. The Biochemical Journal. 1993;292(3):839-844. | ||
| + | |||
| + | <div id="Data"></div>[5] Saccharomyces Genome Database [Online]. Stanford: National Human Genome Research Institute; 1997 [updated 2012 Feb 9; cited 2012 May 13]. Available from: http://www.yeastgenome.org/. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
</div> | </div> | ||
{{Team:Chalmers-Gothenburg/footer}} | {{Team:Chalmers-Gothenburg/footer}} | ||
Revision as of 15:09, 3 July 2012
Notation! This page is under construction --Annaru 10:09, 3 July 2012 (CDT)
Contents |
Methods
The work required for the construction of the biosensor is divided into three main tasks: (1) expression of the LH/CG receptor, (2) knock-out of the CWP2 gene encoding a cell wall mannoprotein and (3) expression of indigo synthesizing enzymes tryptophanase and monooxygenase. These tasks will be performed in parallel and in the end, the different parts will be combined into one system. The three main tasks are described in detail in the following sections.
Expression of the LHCGR gene in yeast strain IMFD-73
The first task is to express the LH/CG receptor in yeast. The receptor contains several amino acid residues that are essential for efficient ligand binding, both in the extracellular and in the transmembrane domains. In addition, the C-terminus and the intracellular loop III are considered to bind to the associated Gα protein of the receptor, and thereby constitute an important part of the signal transduction [1]. In order to ensure correct conformational change following ligand binding and to guarantee a proper signal transduction, the full-length LH/CG receptor will be expressed. The LHCGR gene will be inserted into the yeast strain IMFD-73 and be placed under the control of a strong, constitutive promoter to ensure constant expression. To enhance translation efficiency, a Kozak sequence (AAAACA) will also be inserted upstream of the LHCGR gene. In addition, a signal peptide will be added to the N-terminus of the receptor sequence to ensure transportation to the yeast cell membrane.
Previous research does not provide general guidelines for which signal peptide should be used when expressing human GPCRs in yeast. In this project, both the native signal of the LH/CG receptor, consisting of its 22 first amino acids [2], and a yeast signal peptide will be tested separately. The 5' end of the LH/CG receptor, which will be used as the native signal peptide, is originally glycosylated and glycosylation in yeast differs significantly from the one in human [3]. The native signal will, however, be tested even though it is not sure if glycosylation of these amino acid residues is required for correct transportation of the receptor to the membrane [4]. In order to test a yeast signal, the idea was to fuse the signal peptide of Ste2, the receptor to be replaced, with the LH/CG receptor N-terminus. However, Ste2 lacks an identified signal sequence [5]. Instead, the signal peptide from Ste3, the MATα type pheromone receptor, will be used. It consists of the first 25 amino acid residues of the receptor [5].
Plasmid construction
The LHCGR gene construct will be ordered in a vector. Figure 1 provides a schematic illustration of this construct. The signal peptide of Ste3 and a Kozak sequence will be fused with a Kozak sequence at the 5' site of the native signal and the gene. In addition, NotI and one PacI restriction sites will be added as indicated in Figure 1.
Figure 1: Schematic illustration of the LHCGR gene construct. Using this construct enables expression of the LHCGR gene with each signal peptides separately. For the expression of the gene with the native signal, the vector can be digested directly with NotI and PacI. When testing the expression with the yeast signal, PCR can be run with 5' phosphorylated primers indicated by arrows in order to exclude the native signal and its associated Kozak sequence and PacI restriction site. The resulting linear fragment can be religated.
The aim of using this particular construct is to enable expression of the LHCGR gene with both signal peptides separately. For the expression with the native signal, the vector containing the LHCGR gene will be digested directly with NotI and PacI restriction enzymes. The fragments will be purified on a gel and then ligated into the corresponding sites of the expression plasmid pSP-GM1 with bidirectional promoters TEF1 and PGK1, a URA3 marker gene and an ampicillin resistance gene. For expression of the LHCGR with the Ste3 signal peptide, PCR will be run with 5' phosphorylated primers indicated with arrows in Figure 1 (lhcgr-rev and lhcgr-fw). This will result in a linear fragment that lacks the native signal and its associated NotI site and Kozak sequence. This fragment will be purified on a gel, religated and amplified in E. coli. After that, it will be digested with NotI and PacI restriction enzymes, column purified and then ligated into the expression plasmid pSP-GM1.
In both cases, the gene will be set under the control of the strong and constitutive TEF1 promoter. The resulting plasmids will be named pLHCGRn (native signal peptide) and pLHCGRy (yeast signal peptide). Figure 2 shows a map of the plasmid pLHCGRn. pLHCGRy is similar to pLHCGRn and only differs in length of the LHCGR gene construct, which is 9 bp longer in the pLHCGRy plasmid. For amplification, E. coli strain DH5α will be transformed separately with the two plasmids and an empty control plasmid by heatshocking. The cells will be cultivated in ampicillin-containing LB medium.
Figure 2: Plasmid map of pLHCGRn derived from pSP-GM1. The plasmid contains a yeast origin of replication, 2μ ori, and a yeast marker, URA3. The parts that are important for transforming E. coli are the ampicillin resistance gene, AmpR and the E. coli origin of replication, pUC ori. The LHCGR gene construct with the yeast signal peptide is anked by NotI and PacI. P TEF1 is a strong, constitutive promoter and T ADH1 is a terminator.
References
[1] Ryu KS, Ji L, Chang L, Ji TH. Molecular mechanism of LH/CG-receptor activation. Molecular and Cellular Endocrinology. 1996;125(1-2):93-100. [2] Dufau ML. The hormone luteininizing hormone receptor. Annual review of Physiology. 1998;60(1):461-496. [3] Hamilton SR, Gerngross TU. Glycosylation engineering in yeast: the advent of fully humanized yeast. Current Opinion in Biotechnology. 2007;18(5):387-392. [4] Petaja-Repo UE, Merz WE, Rajaniemi HJ. Significance of the carbohydrate moiety of the rat ovarian luteinizing-hormone/chorionic-gonadotropin receptor for ligand-binding specificity and signal transduction. The Biochemical Journal. 1993;292(3):839-844. [5] Saccharomyces Genome Database [Online]. Stanford: National Human Genome Research Institute; 1997 [updated 2012 Feb 9; cited 2012 May 13]. Available from: http://www.yeastgenome.org/.
 "
"