Team:Chalmers-Gothenburg/Biodetection of hCG
From 2012.igem.org
m (→Our Design: How the Biosensor works) |
m (→Our Idea: Biodetection of the hCG Hormone) |
||
| Line 2: | Line 2: | ||
<div align="justify" style="width:auto; margin-left:50px; margin-right:50px;"> | <div align="justify" style="width:auto; margin-left:50px; margin-right:50px;"> | ||
== Our Idea: Biodetection of the hCG Hormone== | == Our Idea: Biodetection of the hCG Hormone== | ||
| - | Our project idea is to develop a biosensor for human chornionic gonadotropin (hCG), which is a hormone that is produced in the body during pregnancy. Consequently, by using this new technique for detecting the hCG hormone, we would like to create a biodegradable pregnancy test kit. To construct the biosensor, we will use ''Saccharomyces cerevisiae'' and replace its endogenous G protein-coupled | + | Our project idea is to develop a biosensor for human chornionic gonadotropin (hCG), which is a hormone that is produced in the body during pregnancy. Consequently, by using this new technique for detecting the hCG hormone, we would like to create a biodegradable pregnancy test kit. To construct the biosensor, we will use ''Saccharomyces cerevisiae'' and replace its endogenous G protein-coupled receptor, Ste2, with the human luteinizing hormone receptor (LH/CG), which is the receptor that binds to hCG with high affinity. For an output signal, we will also attempt to enable ''Saccharomyces cerevisiae'' to produce indigo in response to detecting hCG. This will be done by coupling our receptor and genes needed for indigo production, with the yeast pheromone pathway. |
There are several novel aspects of this project. The receptor that we will utilize in our biosensor has never been expressed in yeast before. In addition, production of indigo has never successfully been carried out in yeast earlier and we hope that indigo can serve as a new and efficient reporter. We would also like to illustrate the usefulness of exploiting the pheromone pathway in yeast when constructing a biosensor. Naturally, we also hope that our new technique for detecting hCG could contribute to the development of a cheap, biodegradable and reusable pregnancy test. | There are several novel aspects of this project. The receptor that we will utilize in our biosensor has never been expressed in yeast before. In addition, production of indigo has never successfully been carried out in yeast earlier and we hope that indigo can serve as a new and efficient reporter. We would also like to illustrate the usefulness of exploiting the pheromone pathway in yeast when constructing a biosensor. Naturally, we also hope that our new technique for detecting hCG could contribute to the development of a cheap, biodegradable and reusable pregnancy test. | ||
Revision as of 13:01, 6 July 2012
Our Idea: Biodetection of the hCG Hormone
Our project idea is to develop a biosensor for human chornionic gonadotropin (hCG), which is a hormone that is produced in the body during pregnancy. Consequently, by using this new technique for detecting the hCG hormone, we would like to create a biodegradable pregnancy test kit. To construct the biosensor, we will use Saccharomyces cerevisiae and replace its endogenous G protein-coupled receptor, Ste2, with the human luteinizing hormone receptor (LH/CG), which is the receptor that binds to hCG with high affinity. For an output signal, we will also attempt to enable Saccharomyces cerevisiae to produce indigo in response to detecting hCG. This will be done by coupling our receptor and genes needed for indigo production, with the yeast pheromone pathway.
There are several novel aspects of this project. The receptor that we will utilize in our biosensor has never been expressed in yeast before. In addition, production of indigo has never successfully been carried out in yeast earlier and we hope that indigo can serve as a new and efficient reporter. We would also like to illustrate the usefulness of exploiting the pheromone pathway in yeast when constructing a biosensor. Naturally, we also hope that our new technique for detecting hCG could contribute to the development of a cheap, biodegradable and reusable pregnancy test.
Our Design: How the Biosensor Works
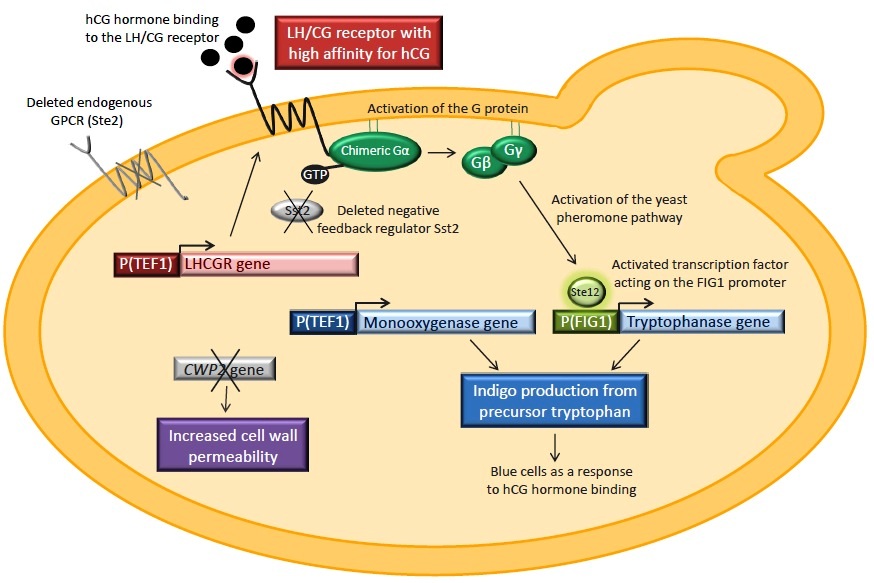
The yeast strain that will be used is provided by the Kondo group from Kobe University in Japan. It has the endogenous GPCR Ste2 deleted in order to prevent it from isolating G proteins in inactive complexes. Sst2, a negative feedback regulator of the pheromone pathway, is also knocked out. The strain contains a yeast/human chimeric Gα subunit which should ensure the interaction of the human GPCR with the yeast pheromone pathway. Binding of hCG to the LH/CG receptor will result in the activation of the chimeric G protein, the activation of the pheromone pathway and consequently the expression of indigo synthesizing enzymes and blue cells.
Our project includes the constitutive expression of the human luteinizing hormone receptor (LHCGR). Two signal peptides will be tested when expressing the receptor, the native signal and a yeast membrane protein signal. The genes tnaA and fmo, encoding tryptophanase and flavin-containing monooxygenase respectively, will also be introduced into the yeast strain. These enzymes catalyze the conversion of tryptophan to indigo. TnaA will be coupled with the pheromone pathway by setting it under the control of the pheromone-induced FIG1 promoter whereas the fmo will be expressed constitutively. Another part of the project is the deletion of the CWP2 gene, encoding a cell wall mannoprotein, in order to increase the cell wall permeability. This is done in oder to enhance the chances of the hCG hormone to pass the yeast cell wall and to bind to the receptor.
An overview of the biological construct that should serve as a biosensor for hCG. The LH/CG receptor is expressed in yeast. STE2 and SST2 are deleted and the strain expresses a human/yeast chimeric Gα subunit. The tryptophanase gene is under the control of the pheromone-induced FIG1 promoter whereas the monooxygenase is expressed constitutively. CWP2 is removed in order to increase the cell wall permeability.
Expression of LH/CG receptor
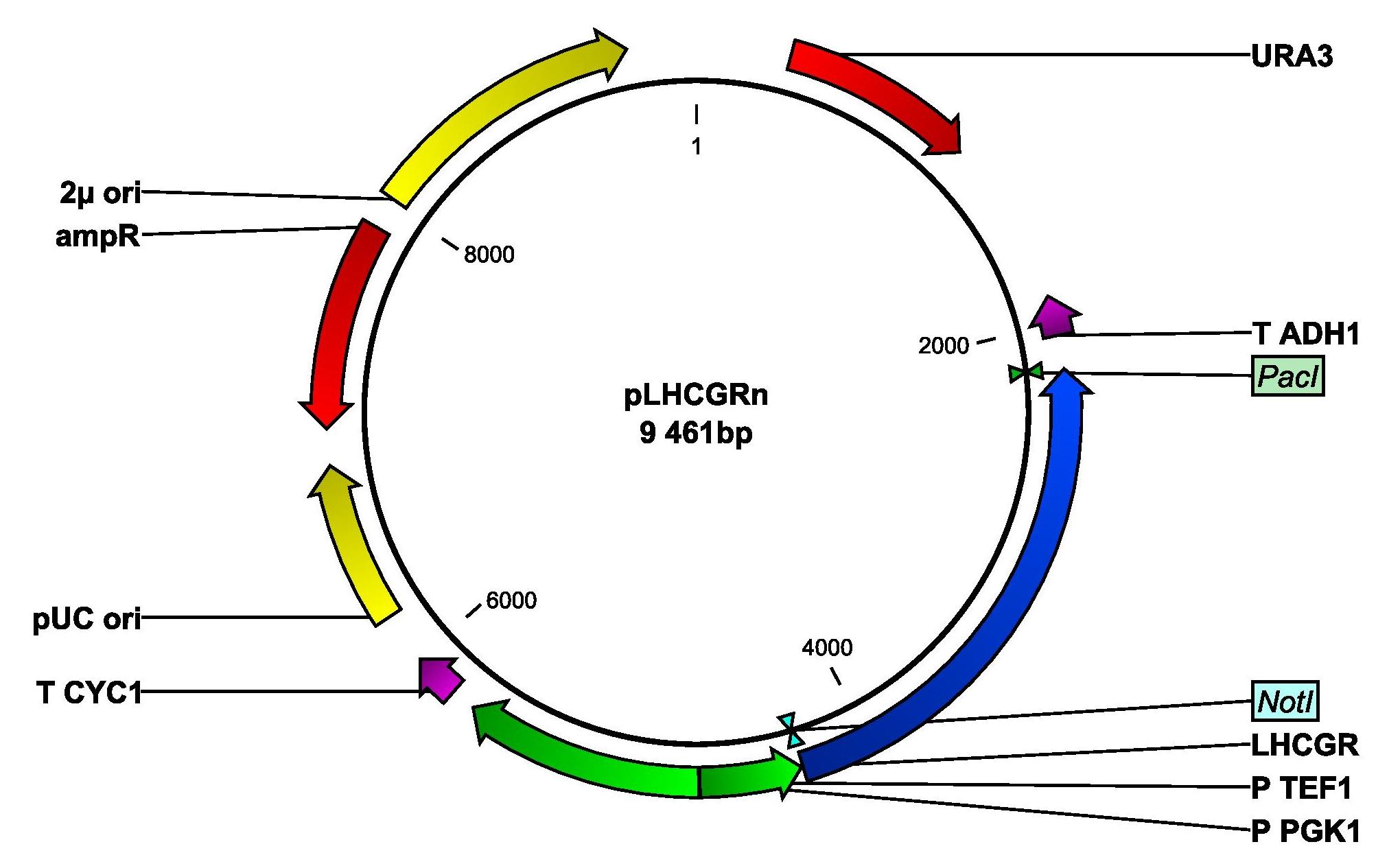
We will express the The LH/CG receptor with either the native or a yeast signal peptide. The map of the plasmid that we will construct and transform yeast with can be seen below.
The map of the plasmid that we will insert into yeast. The LHCGR gene is set under the control of the strong, constitutive promoter PTEF1.
Expression of indigo synthesizing enzymes
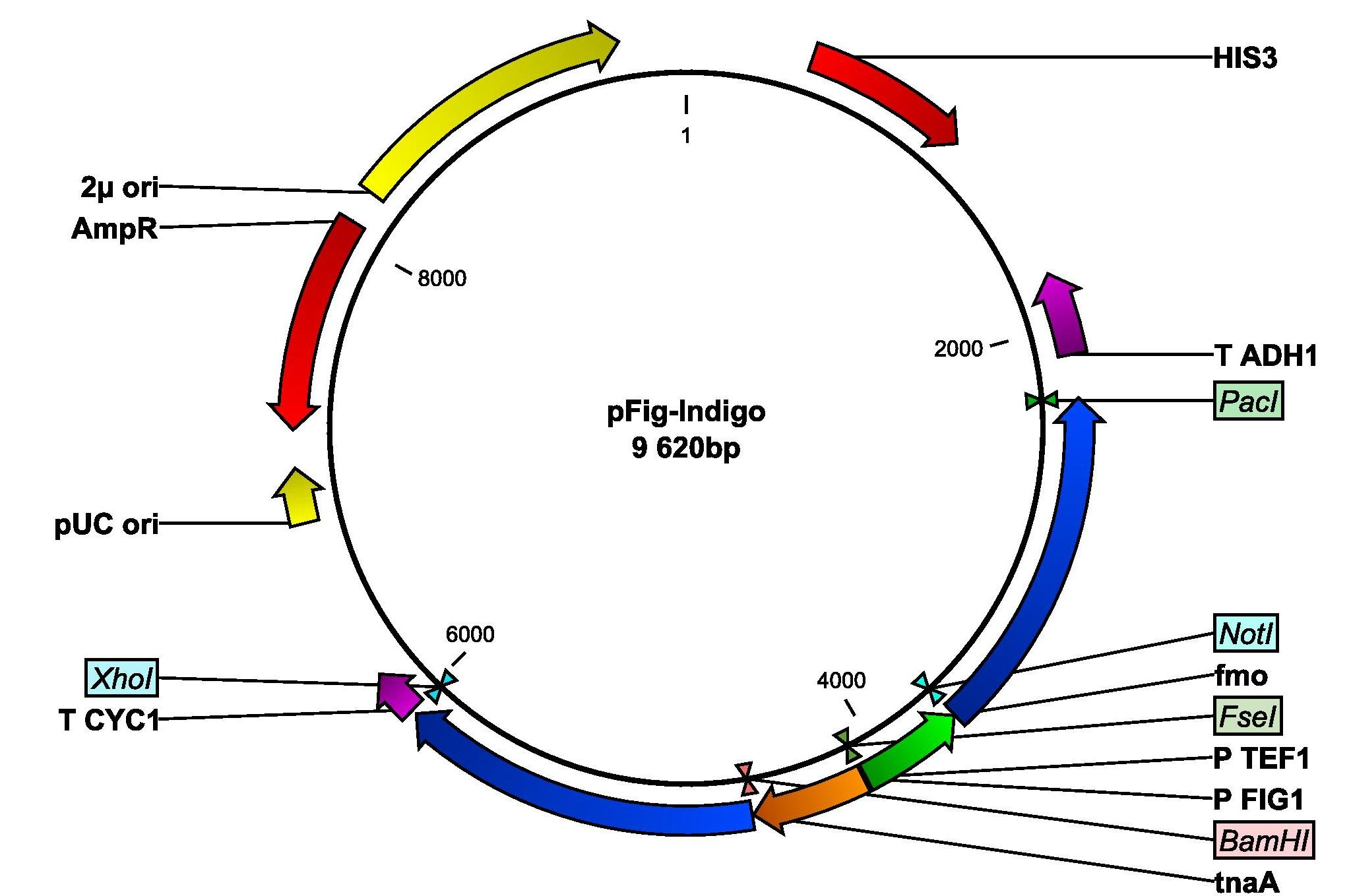
We will insert the two genes fmo and tnaA into yeast with the following plasmid. The tnaA gene is amplified from E.colis genomic DNA and is set unter the control of the pheromone-induced FIG1 promoter. The fmo gene, kindly provided by Si Wouk Kim from Chosun University in South Korea, is set under the control of the constitutive TEF1 promoter.
Plasmid map of the PFigIndigo that will be inserted into yeast. The plasmid contains the genes fmo and tnaA whereof the tnaA genes set under the control of the pheromone-induced FIG1 promoter and thus coupled with the pathway. The fmo gene is set under the control of the TEF1 promoter.
Deletion of the CWP2 gene
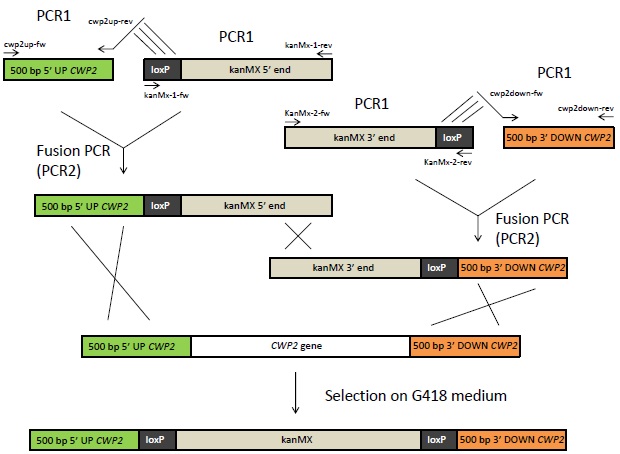
The yeast cell wall limits the permeability and large molecules will not easily pass through it [1]. Hence, if a ligand of a heterologously expressed GPCR is larger than 70-80 amino acid residues, there is risk that it will not be able to activate the receptor [1]. The hCG hormone, the ligand in this project, has two subunits consisting of 92 and 145 amino acids respectively [2] and this could constitute a problem for the hCG biosensor. To solve this problem, we delete the CWP2 gene, which encodes a cell wall mannoprotein. This will probably increase the cell wall permeability and hopefully allow hCG to pass through the cell wall. The gene deletion is attempted by using the Bipartite method, which is outlined below.
Overview of the Bipartite method. 500 bp of the up- and downstream sequence of the CWP2 gene will be amplified from genomic DNA using primers with 5' extensions that are complement to the loxP sites as indicated with lines above. The 5' and 3' end of the kanMX (kanamycin resistance) gene flanked to loxP sites, available in vectors, will also be amplified by PCR. The fragments from these PCR1 reactions will be fused by Fusion PCR. The resulting fragments of this reaction can then be inserted into yeast. Homologous recombination will lead to the exchange of CWP2 with kanMX in the yeast's genome. The cells can be selected on G418 medium.
References
[1] Ladds G, Goddard A, Davey J. Functional analysis of heterologous GPCR signaling pathways in yeast. Trends in Biotechnology. 2005;23(7):367-373. [2] Cole LA. hCG. Five independent molecules. Clinica chimica acta; international journal of clinical chemistry. 2012;411(1-2):48-65.
 "
"