Team:USTC-China/methods
From 2012.igem.org
(img) |
(img) |
||
| Line 269: | Line 269: | ||
own transcription. </p><br> | own transcription. </p><br> | ||
<div class="imgholder1" align="left" style="float:left;clear:both;width:800px;height:auto;"> | <div class="imgholder1" align="left" style="float:left;clear:both;width:800px;height:auto;"> | ||
| - | <a href="https://static.igem.org/mediawiki/2012/ | + | <a href="https://static.igem.org/mediawiki/2012/a/ac/The_bacteriophage_l_CI_protein_finds_an_asymmetric_solution_%28b%29.png"><img src="https://static.igem.org/mediawiki/2012/a/ac/The_bacteriophage_l_CI_protein_finds_an_asymmetric_solution_%28b%29.png" alt="Bacteriophage_lambda_genome_total" style="clear:both;width:800px;height:287px;"></a><br><p><small align="center">CI dimers bound cooperatively to adjacent operator sites in OR and OL. The CI dimers are shown in blue. Each subunit of the dimer consists of an N-terminal DNA-binding domain (N), a C-terminal oligomerization domain(C), and a linker region (black) connecting the two. The dimer pair bound cooperatively at OR1 and OR2 represses transcription from PR and the dimer bound at OR2 also activates transcription from PRM. The dimer pair bound cooperatively at OL1 and |
OL2 represses transcription from PL.</small></p> | OL2 represses transcription from PL.</small></p> | ||
| Line 279: | Line 279: | ||
stable level.</p> | stable level.</p> | ||
<div class="imgholder1" align="left" style="clear:both;width:546px;height:auto;"> | <div class="imgholder1" align="left" style="clear:both;width:546px;height:auto;"> | ||
| - | <a href="https://static.igem.org/mediawiki/2012/ | + | <a href="https://static.igem.org/mediawiki/2012/8/88/The_bacteriophage_l_CI_protein_finds_an_asymmetric_solution_%28c%29.png"><img |
| - | src="https://static.igem.org/mediawiki/2012/ | + | src="https://static.igem.org/mediawiki/2012/8/88/The_bacteriophage_l_CI_protein_finds_an_asymmetric_solution_%28c%29.png" |
alt="Bacteriophage_lambda_genome_total" style="clear:both;width:546px;height:223px;"></a><br><p><small align="center">Higher-order looped | alt="Bacteriophage_lambda_genome_total" style="clear:both;width:546px;height:223px;"></a><br><p><small align="center">Higher-order looped | ||
Revision as of 13:21, 26 September 2012
METHODS
Overview of our project
In fermentation production, the loss results from the bacteria's being infected by the phage is striking. Because the phage infect intensely and spread rapidly, once the bacteria are infected, it is very difficult to eradicate the phages and the lysogens thoroughly. If the fermentation broth is unluckily polluted severely, the factory will have to stop production totally, and massive loss in profit happens afterward. For this reason, it is quite important to prevent the invasion of the phages. (see the background)
To help solve this problem, this year, the USTC iGEMers designed a gene circuit for bacteria which can both detect and defend against the phages.
Generally, defence can be classified as active defence and passive defence. If we make our engineered bacteria defend actively, they will expend large amounts of energy fighting against the phages even in an environment without phages. If so, the production of fermented products will be affected. For this reason, we successfully design our bacteria which can detect the infection of the phages. Only when the phage infects the host, the host will start its resistance against the phage.
We kill the phage by means of making the host suicide. Provided that the host can suicide fast enough and die before newly assembled phages become mature, the new waves of infection will be successfully avoided. In this case, the damage the phages can cause will be much lower. What's more, our engineered bacteria can efficiently kill the lysogenic bacteria which hide in the colonies. This will guarantee the colony's safety from the origin.
However, the phage is still possible to release plenty of mature phages before the host suicides. What's worse, the circumstance in the fermentation tanks is very beneficial for bacteria to grow. And when a lysogenic bacterium lives in a fertile circumstance, the phage tend to transform into lytic life cycle.(see the "Lytic or lysogenic?" in the background page). In order to prevent the phage from reproducing and releasing large amounts of newly assembled phages, we must decrease the possibility for the phage to turn into the lytic life cycle and make it stay at lysogenic life cycle. In this case, we can win plenty of time for this bacterium to defeat the phage.
At the same time, to keep the safety of the colony better, we use the quorum sensing system to make the bacteria around the host start to defend. But due to the complexity of the E.coli's membrane, we failed to find methods to prevent the phages from invading into the bacteria. Also because of deficiency of time, we did not complete the quorum sensing system.
Here is the gene circuit we designed.

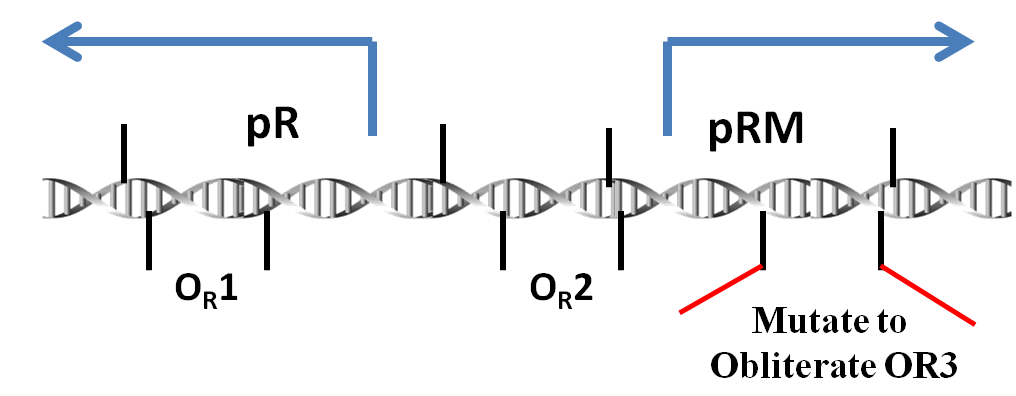
We use the promoter pRM(originally from the genome of lambda phage) to detect whether there are phages invading the bacteria. If a phage infects a bacterium, the protein CI which the phage produces will bind the site OR1 and OR2. Thus, the pRM will be activated and the products downstream the pRM will be expressed. What's more, the protein Cro cannot inactivate the pRM since our pRM has been modified. There are only sites OR1 and OR2 but no OR3. All the genes we use for resistance to phages are downstream.
We use the lysis as a suicide gene to kill the lambda phage and its host together. The lysis protein causes modifications of inner membrane. Then it activates OmpLA to destroy the outer membrane, which will result in cell death. When the lysis protein reaches a critical concentration, the death effect occur. If the phage cannot turn into lytic life cycle or newly assembled phages are still immature, no waves of infection will occur afterwards.
We use the antisense RNA to prevent the phage from turning into lytic life cycle. Downstream of the pRM there is a cro gene which is ligated reversely without a RBS ahead of it. This gene can express antisense RNA (we call it anticro) which can complementarily bind with the mRNA of cro. If the mRNA is bound by the anticro, it cannot translate the protein Cro and will degrade soon. In this case, the amount of Cro will decrease and repressor CI will make lambda phage stay at the lysogenic life cycle.

We use the modified promoter pRM to sense the phage’s infection and initiate the defence. The lysis gene which can make bacteria lyse is installed in our circuit.To win more time for lysis to function well, we design antisense RNA to prevent the phage from turning into lytic life cycle.
pRM
Promoter pRM is a component of the genome of lambda phage. To be more exact, it plays an important role in the toggle switch in lambda phage.
In lambda phage, pRM initializes the transcription leftwards. There are three protein binding regions OR1, OR2, OR3 and a promoter pR (which initiates transcription in opposite direction) around the pRM. OR1, OR2, OR3 can be bound by protein CI dimmers(CI2) and Cro dimmers(Cro2), but the affinities of these three region of interacting with CI2 or Cro2 are different from each other. The sequences of these three binding sites and the two promoter pRM and pR are overlapped with each other(see the picture below), thus the transcription from these two promoters will be affected if the CI2 or Cro2 binds The sites of these three regions on DNA are shown in the picture below.
There is only one gene downstream of the pRM, namely cI and lysogeny is maintained solely by cI. cI represses transcription from pL and pR while upregulating and controlling its own expression from pRM. It is therefore the only protein expressed by lysogenic phage. CI2 binds most favorably to OR1; binding here inhibits transcription from pR. The binding of CI2 to OR1 greatly increases the affinity of the binding of cI to OR2, and this happens almost immediately after OR1 binding. This activates transcription in the other direction from pRM, as the N terminal domain of cI on OR2 tightens the binding of RNA polymerase to pRM and hence cI stimulates its own transcription.
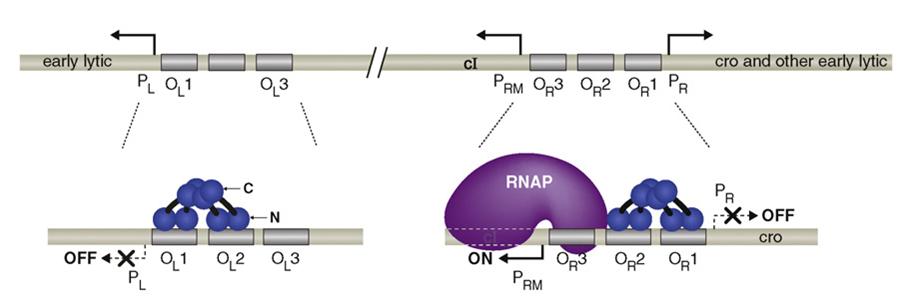
CI dimers bound cooperatively to adjacent operator sites in OR and OL. The CI dimers are shown in blue. Each subunit of the dimer consists of an N-terminal DNA-binding domain (N), a C-terminal oligomerization domain(C), and a linker region (black) connecting the two. The dimer pair bound cooperatively at OR1 and OR2 represses transcription from PR and the dimer bound at OR2 also activates transcription from PRM. The dimer pair bound cooperatively at OL1 and OL2 represses transcription from PL.
When it is present at a much higher concentration, it also binds to OR3, inhibiting transcription from pRM, thus regulating its own levels in a negative feedback loop. Thus, when the phage stays at lysogenic life cycle, the concentration of CI can maintain at a stable level.

Higher-order looped complex that facilitates negative autoregulation. When CI2 binds at OR3, CI2 represses transcription from pRM (negative autoregulation); however, in a lysogen OL3 and OR3 are only partially occupied.
Function in our project
Since the lambda phage expresses protein CI after infecting and the pRM will be activated by the dimer CI2, we choose this promoter to confer our engineered bacteria the ability of detecting the phage's infection.
In order to prevent Cro and overexpressed CI from repressing the pRM, we choose a modified pRM in partsregistry. In wild type lambda phage, the OR3 site (-27 to -11) reads "tatcccttgcggtgata" on the sense strand of DNA. In the modified pRM, the 4th through 10th nt of OR3 were replaced by the 7 nt between OR2 and OR1, "aaatagt" (-57 to -51), which in effect mutates 5 nt of the promoter. These nt were chosen to be about halfway between the -10 and -35 boxes of the promoter. Furthermore, since these nt already act as a spacer in wild-type phage, it is hoped they will not create undesired interactions.
Antisense RNA
Antisense RNA (asRNA) is a single-stranded RNA that is complementary to a target mRNA. Antisense RNA in vivo can inhibit translation of a complementary mRNA by base pairing to it and physically obstructing the translation.[1] For many years, noncoding RNAs produced in eukaryotes have been a research hotspot. Until very recently, due to the accumulating evidence from transcriptome studies suggesting that extensive antisense transcription also occurs in bacteria, people realized that asRNA could be a significant level of regulation on gene expression. However, the functions asRNA can realize have only partially been discovered and the mechanisms of asRNAs' interactions with target mRNAs are remain hypothetic or unclear. What asRNAs can do and how they do that? Here let's take an overview.
What can asRNA do?
The direct function of asRNA is repressing the translation of the target mRNA. However, since one kind of asRNA is only produced under certain conditions, the asRNA is actually an efficient approach prokaryotes can take to regulate gene expression and hence modulate the metabolism.
How they do that?
In many different cases can asRNA regulates gene expression. The mechanisms of asRNA action in bacteria can generally classified as below:
1. Transcription interference
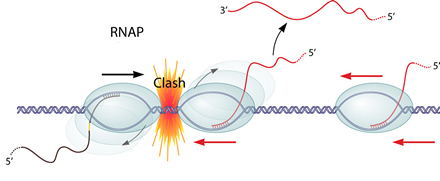
Transcription interference occurs when transcription from a promoter is suppressed by another one present in cis. If two RNAPs(RNA polymerase) initiate transcription from two promoters in opposite direction, these two RNAPs will collide and may dissociate from the transcription complex. What's more, if one promoter is far too stronger than the other one, the RNAP coming from the "aggressive" promoter will prevent the formation of the transcription complex at the "sensitive" promoter.

the RNAP coming from the "aggressive" promoter will prevent the formation of the transcription complex at the "sensitive" promoter.
2. Transcription attenuation
In a few cases, by base paring, the asRNA can induce the formation of a terminator structure in the target mRNA. Thus, the target mRNA is prematurely terminated. The classical example is the Shigella flexneri virulence gene icsA (virG), which is controlled by the asRNA RnaG.

The 5' structure of the target mRNA resembles an antiterminator structure (2a), and the full-length mRNA could be transcribed (3a). When asRNA is present, it forms a heteroduplex with the growing mRNA. This inhibits the formation of the antiterminator structure, and a terminator hairpin is formed (2b). Subsequently, transcription is attenuated, and the immature mRNA is released. (3b).
3. RNA cleavage
The interaction of an asRNA with its target RNA alters the secondary structure of both two RNAs and results in duplex (double- stranded RNA [dsRNA]) formation. Those structural changes influence the stability and half-life of the RNAs. In some cases, duplex formation results in the rapid and complete degradation of both RNAs. The endoribonucleases involved in degradation are listed below:
1) RNasr III, which cleaves dsRNA. The duplex formed by the asRNA and its target is the ideal substrate for RNase III. (eg. copT- copA and hok-sok plasmid-based sense-asRNA)
2) RNase E, which cleaves ssRNA. It is conceivable that the 5' -end of an antisense RNA could provide a monophosphate which might stimulate RNase E activity.

4. Translation block
Many asRNAs base pair with the RBS (ribosome binding site) of their target mRNAs thereby preventing ribosome binding and protein translation. Antisense RNAs for which complimentarity extends into the 5' UTR (untranslated region) of the mRNA target may act by a similar mechanism. What's more, the binding of the asRNA directly or indirectly triggers the enhanced degradation of the target mRNA.
Generally, antisense RNA still lack effective design, biological activity, and efficient route of administration.
Function in our project
We use the asRNA of cro, namely anticro, to repress the expression of the protein Cro. In this way, we prevent the lambda phage
from turning into lytic life cycle and win more time for protein lysis to function well. The success of our system clearly relies on
the ability of the anticro to effectively and specifically repress the expression of the protein Cro.
In order to test the ability of the anticro to repress the expression of cro, we construct a fusion protein of cro and gfp with a linker (GGSG,[BBa_K648005]) (see our description of crogfp[BBa_K741006] on the partsregistry.org). In this way, we can examine the efficiency of the anticro by means of measuring the fluorescence intensity of the crogfp.
Lysis
Introduction
Colicin lysis proteins are small lipoproteins of 27 to 35 amino acids whose sequences exhibit a high degree of similarity, which is suggestive of a common origin.The main function of the protein is to promote colicin release.Concomitantly, they provoke quasilysis;modifications of the cell envelope;activation of OmpLA,the outer membrane phospholipase A;and death of the producing cell.
Function

The suicide process induced by lysis gene. (a)The activated lysis gene express the lysis protein. (b)The lysis protein accumulate on the inner membrane and when the lysis protein reach a critical concentration ,the protein cause modification of the inner membrane.(c)The lysis protein pass through the modified inner membrane and activate the OMPLA between the two membranes.Then the OMPLA destroy the outer membrane. (d)The host cell dies due to the damage of the outer membrane.

A contrast of normal E.coli cells and lysed E.coli cells.(a)Normal cells without induction.(b)The cells are induced to express lysis protein.
Though the precise mechanism of the lysis process is still unknown,some researches have made progress. Two processes occur during the cell death.First, the lysis protein causes modifications of inner membrane. Then activates OmpLA to destroy the outer membrane, which will result in cell death. When the lysis protein reaches a critical concentration,the death effect occur.
Function in our project
In our project,the lysis gene is applied as a suicide gene on the basis of the cell-death effect.
We plan to block the spread of bacteriophage with this property. Once the bacteriophage invade the host cell, the lysis gene is simulated to express. Soon the cell suicides and the lambda phage can no longer take advantages of the enzymes and the energy of the host to replicate. At that time, the new generations of bacteriophage are immature. As we know,immature bacteriophage has no invasion activity. Thus, the growth of bacteriophage is resisted.
Reference
 "
"