Contents |
Why?
From our work on LOV2 photoactivation we should be able to predict the percentage of LovTAP-VP16 proteins that are photoactivated depending on the irradiance applied.
- Can we then predict the occupancy of TrpO binding sites in our reporter plasmid by LovTAP-VP16 dimers?
- And the difference in expression levels between an irradiated cell culture a one in the dark?
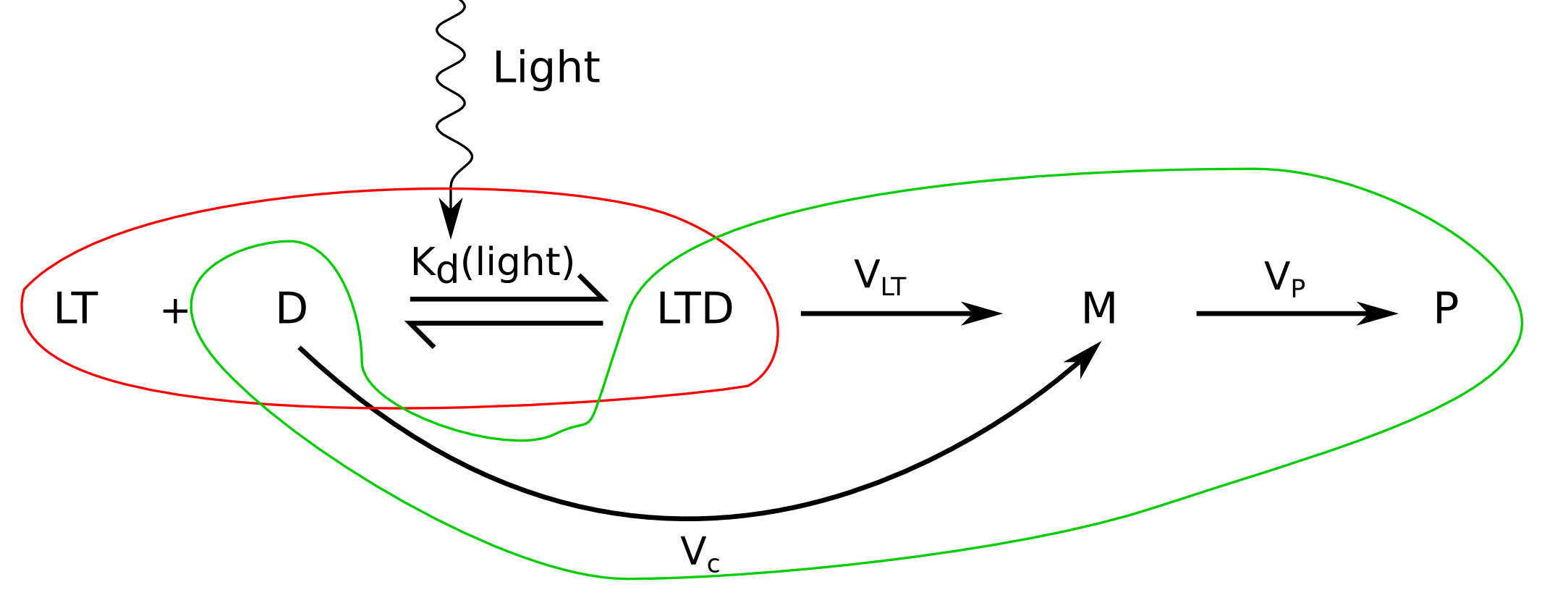
What?
Well, things start to get complicated, since the number of unknowns is huge in this problem, because it's affected by the transient transfection process, the cell's metabolism, the interaction of plasmids with the genomic DNA, the interaction of VP16 with the transcriptional cofactors and lots more.
We can split this into two parts:
- Estimation of the proportion of active plasmids that will be bound to LovTAP-VP16.
We will assume the same binding affinity ratio (light/dark) as Strickland et al (2008) found for LovTAP at 25ºC.
Yang et al (1996) our TrpO sequence might host 1 or 2 LovTAP-VP16 dimers.
How?
Photoactivation level
We will then assume that we know the proportion of activated LovTAP-VP16 monomers, since we have calculated it in this section. But LovTAP-VP16 has to dimerize to be able to bind DNA. To illustrate the importance of this, let's give an example: imagine a solution with LovTAP-VP16 where 50% of the monomers are activated. Assuming that all of them form part of a dimer, we get that 25% of the dimers are inactive, 25% active and 50% half active. The average binding affinity will mostly depend on the affinity of the half active half inactive dimers. In the section on the 3D protein structure of LovTAP-VP16 we conclude that it is reasonable to think that a half activated dimer will have a DNA activity somewhere between a fully activated dimer and a deactivated one. This means that we can take the monomer activation level to approximate the dimer activation level.
92% of photoactive LOV2 has it's Jα-helix undocked (Yao et al, 2008).
Eitoku et al (2005) say it takes 2 ms for the J&alpha-helix to undock.
 "
"