Team:Shenzhen/Result/YAO.Genome
From 2012.igem.org
(Difference between revisions)
| Line 81: | Line 81: | ||
<div class="context"> | <div class="context"> | ||
<h5>YAO.Genome Backbone</h5> | <h5>YAO.Genome Backbone</h5> | ||
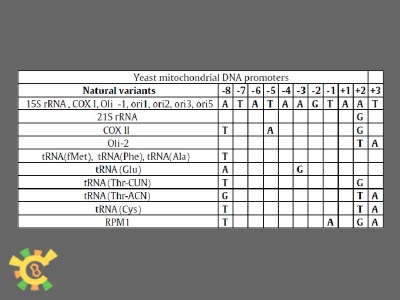
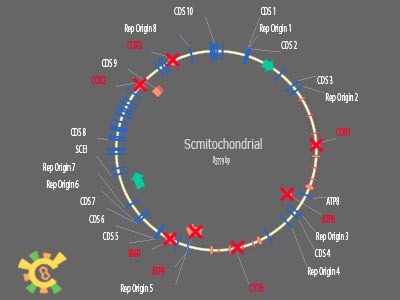
| - | <ul><p> | + | <ul><p> We also minimized the mitochondrial genome, with the annotation database. Deleting all know coding sequences, reserving replication related, translational and transcriptional elements, and non-studied sequences, to be the YAO.Genome backbone.</p> |
[[File:result_yao_genome_p9.jpg]] | [[File:result_yao_genome_p9.jpg]] | ||
| - | <p>Figure 8. YAO.Genome | + | <p>Figure 8. YAO.Genome backbone illustration.</p></ul> |
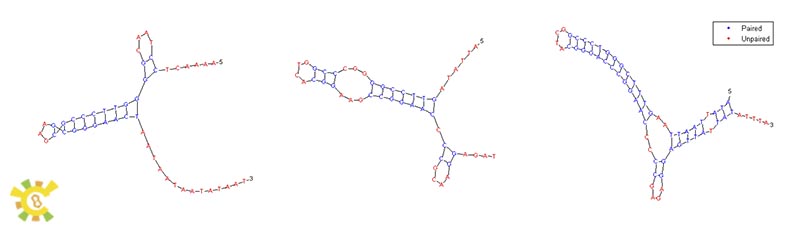
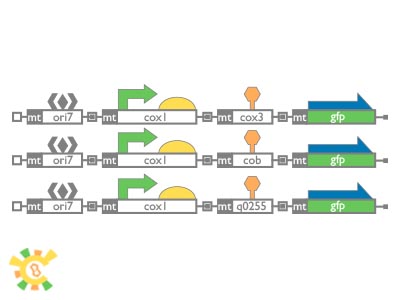
| - | </ul> | + | <ul><p> This figure below shows our image of our YAO. Genome which will have all features as mentioned above. Segments with different colors represent different genes. These genes constructed different systems. Different systems in YAO. Genome will provide clues to improve producing efficiency, purity, safety and to further research about nucleus and mitochondria co-regulation. Maybe we can achieve it in the near future.</p> |
| + | [[File:result_yao_genome_p10.jpg]] | ||
| + | <p>Figure 9. YAO.Genome imagination.</p></ul> | ||
| + | |||
</div> | </div> | ||
</div> | </div> | ||
{{:Team:Shenzhen/Temp/gallery.htm}} | {{:Team:Shenzhen/Temp/gallery.htm}} | ||
Revision as of 04:18, 25 September 2012
 "
"