|
|
| Line 244: |
Line 244: |
| | | | |
| | Also, this week we tried to redesign our project. Our first task is to continue with our attempts to make a biobrick with the spider silk monomer ADF-3. We ordered the primers that we need to do that. | | Also, this week we tried to redesign our project. Our first task is to continue with our attempts to make a biobrick with the spider silk monomer ADF-3. We ordered the primers that we need to do that. |
| | + | |
| | Now we are exploring the possibility of improve the product of the secretion system developed by Christopher Voig in his paper “Engineering the Salmonella type III secretion system to export spider silk monomers”. One idea is to attach a growth factor (e.g. recombinant human BMP-2) to the molecule of ADF-3. | | Now we are exploring the possibility of improve the product of the secretion system developed by Christopher Voig in his paper “Engineering the Salmonella type III secretion system to export spider silk monomers”. One idea is to attach a growth factor (e.g. recombinant human BMP-2) to the molecule of ADF-3. |
| | + | |
| | But the problem with the secretion system developed by Christopher Voig is that it works on salmonella, and we cannot work with such an infectious bacteria. | | But the problem with the secretion system developed by Christopher Voig is that it works on salmonella, and we cannot work with such an infectious bacteria. |
| | | | |
| Line 258: |
Line 260: |
| | | | |
| | Furthermore, we studied carefully the paper “Engineering the Salmonella type III secretion system to export spider silk monomers” and its supplementary information. We realized that each protein secreted with this system comes with a 3x FLAG (DYKDDDDK)3. It is a priority for us to find if someone can share us a little of the anti-FLAG antibody (Sigma Cat# F3165). That way, if we use this secretion system we will be able to quantify the production of our protein of interest. | | Furthermore, we studied carefully the paper “Engineering the Salmonella type III secretion system to export spider silk monomers” and its supplementary information. We realized that each protein secreted with this system comes with a 3x FLAG (DYKDDDDK)3. It is a priority for us to find if someone can share us a little of the anti-FLAG antibody (Sigma Cat# F3165). That way, if we use this secretion system we will be able to quantify the production of our protein of interest. |
| - | But, if we want to use Voigt´s system, we need attenuated salmonellas with their pathogenicity island 1 intact. Something similar to Salmonella Typhimurium SL1344. After some research we found a professor of our University that works with salmonella (Dr. Susan Bueno). But she was out of the country and we could only send her some emails asking for the salmonella. | + | But, if we want to use Voigt´s system, we need attenuated salmonellas with their pathogenicity island 1 intact. |
| | + | |
| | + | Something similar to Salmonella Typhimurium SL1344. After some research we found a professor of our University that works with salmonella (Dr. Susan Bueno). But she was out of the country and we could only send her some emails asking for the salmonella. |
| | | | |
| | | | |
| Line 265: |
Line 269: |
| | Finally the primers necessary for biobricking ADF-3 arrived! | | Finally the primers necessary for biobricking ADF-3 arrived! |
| | And we have got other important material: we talked with Dr. Hugo Olguin, and he is willing to share with us his anti-FLAG antibody. Furthermore, he allowed us to do the western blot in his laboratory. | | And we have got other important material: we talked with Dr. Hugo Olguin, and he is willing to share with us his anti-FLAG antibody. Furthermore, he allowed us to do the western blot in his laboratory. |
| | + | |
| | Additionally, we achieved significant advances in the wiki, especially in the section Human Practices. | | Additionally, we achieved significant advances in the wiki, especially in the section Human Practices. |
| | Unfortunately, we still lack of salmonella. | | Unfortunately, we still lack of salmonella. |
| Line 271: |
Line 276: |
| | | | |
| | After some research we found the promising projects of the Japanese iGEM team HokkaidoU 2010 and 2011. They used the E. Coli (k-12) strain SGSC4024, from Salmonella Genetic Stock Center(SGSC) in University of Calgary, Canada (http://people.ucalgary.ca/~kesander/index.html). This strain carries a pBeloBAC11 vector encoding a genome fragment of Salmonella enterica serovar Typhimurium LT2 which covers the SPI-2 region [B_STM07H21 SGSC4024 1464540~1562427] | | After some research we found the promising projects of the Japanese iGEM team HokkaidoU 2010 and 2011. They used the E. Coli (k-12) strain SGSC4024, from Salmonella Genetic Stock Center(SGSC) in University of Calgary, Canada (http://people.ucalgary.ca/~kesander/index.html). This strain carries a pBeloBAC11 vector encoding a genome fragment of Salmonella enterica serovar Typhimurium LT2 which covers the SPI-2 region [B_STM07H21 SGSC4024 1464540~1562427] |
| | + | |
| | We asked the iGEM team members of HokkaidoU 2011 if the E. Coli (k-12) strain SGSC4024 is capable of secreting proteins to the external environment. But unfortunately it seems that is only capable of injecting molecules into other cells, and that is not what we need. | | We asked the iGEM team members of HokkaidoU 2011 if the E. Coli (k-12) strain SGSC4024 is capable of secreting proteins to the external environment. But unfortunately it seems that is only capable of injecting molecules into other cells, and that is not what we need. |
| | | | |
| - | ==July 30 – August 5, 2012 == | + | In the lab we did PCR to amplify ADF-3 and ADF-3+ (i.e. ADF-3 with the tags included in the plasmid that Christopher Voigt sent to us). Unfortunately the PCR did not work. |
| | + | |
| | + | The next day we tried to amplify the backbone pSB1C3, ADF-3+ and ADF-3 with touch-down PCR. The two first amplicons seemed to be all right. Nevertheless, later we purificated and digested the DNA to verificate if the amplicons corresponded to our parts, but the size sequences resulted to be incoherent. We have to try again. |
| | + | |
| | + | ==July 30 – August 5, 2012== |
| | + | This week we did PCR of the backbone pSB1C3, ADF-3+ and ADF-3 again. It failed again. Are our primers wrong? |
| | + | However, we persisted tirelessly with another PCR of pSB1C3, ADF-3+ and ADF-3, this time with a Tm=59º C. It worked for ADF-3+!! |
| | + | |
| | + | We purified the DNA sequence of ADF-3+. Afterwards we did PCR again, but this time to amplify the backbone of ADF-3 that has the promotor pBAD, and to amplify the vector backbone for ADF-3+. |
| | + | |
| | + | The PCR of the backbone of ADF-3 that has the promotor pBAD looks successful! Later we proceeded with the corresponding DNA purification. |
| | + | |
| | + | Joyfully we proceeded with the Gibson Assembly to join ADF-3 to its backbone with pBAD. We transformed the E. coli, but no colony survived. |
| | | | |
| | ==August 6 – 12, 2012== | | ==August 6 – 12, 2012== |
| | + | |
| | + | We did another PCR, but this time amplified ADF-3 with a Tm=56º C ; The vector backbones of ADF-3+ and ADF-3 with Tm=60º C and touch-down PCR of the vector backbones of ADF-3+ and ADF-3 with T annealing = 54º C. |
| | + | Tragically, none of them worked. |
| | + | |
| | + | The next day we did PCR again with Tm=59ºC of ADF-3 and the vector backbones of ADF-3+ and ADF-3. |
| | + | |
| | + | The PCR of ADF-3 was successful! Later we did the purification of the DNA. |
| | + | |
| | + | Afterwards, we completed the Gibson Assambly of ADF3 with the backbone PSB1C3 and the transformations of the E. coli. |
| | + | The next day finally we had some transformed E. coli! Immediately we proceeded with colony PCR to verify if the transformed bacteria correspond to our designs. Unluckily, the colony PCR was negative (the DNA strand that we obtained had a smaller molecular weight than what we expected). |
| | + | |
| | + | So that we repeated the PCR for the vector backbones of ADF-3 and ADF-3+. The bands were from the right length! Afterwards we proceeded with the Gibson Assembly of ADF-3 with its backbone, and of ADF-3+ with its backbone. Both Gibson Assemblies were positive! Finally we continued with a colony PCR. |
| | | | |
| | ==August 13 – 19, 2012== | | ==August 13 – 19, 2012== |
| | + | |
| | + | We purificated the DNA from the colony PCRs that were successful, and then digested it. Fortunately one digestion of ADF-3 and other of ADF-3 had the correct size. |
| | + | |
| | + | Later we did PCR to amplify the backbones of ADF-3 and ADF-3+ (both with pBAD). But we made some errors and added the wrong primers. |
| | + | |
| | + | The next day we repeated the PCR, and the backbone with pBAD of ADF-3 had the right size. |
| | + | Then we send for sequencing ADF-3 and ADF-3+. Later we realized that we sent the wrong sample of ADF-3!! |
| | + | |
| | + | We also transformed some E. coli with backbone with pBAD of ADF-3. The following day we did colony PCR to these E. coli, but we did not obtain the desired results. Then we cultivated the E. coli transformed with the backbone with pBAD of ADF-3 in 200 mL of LB with 1nM of arabinose. |
| | | | |
| | ==August 20 – 26, 2012== | | ==August 20 – 26, 2012== |
| | + | We did colony PCR of 3 colonies transformed with the backbone with pBAD of ADF-3, but the results were negative. |
| | + | The next day we did colony PCR again, but this time with other method: |
| | + | |
| | + | 1) Put 100 mL of bacteria in eppendorf tubes of 2µl and heat at 95ºC for 10 minutes. |
| | + | |
| | + | 2) Centrifuge at maximum speed for 5 minutes. |
| | + | |
| | + | 3) Do Colony PCR with the supernatant as the template. |
| | + | |
| | + | Unfortunately, the results were negative. |
| | + | |
| | + | And we became the results from the sequencing. ADF-3 is incorrect (as we expected, because we sent the wrong sequence), and ADF-3+ is a bit weird. Bernardo never trusted the PCR band of this sequence anyway… |
| | + | |
| | + | The next day we sent for sequencing ADF-3 (this time the right sequence). Some days later we became the results from this sequencing, and it were negative. Dammit! |
| | + | |
| | + | Finally, we prepared the materials for the community outreach activity for Penta UC, the Talents Development and Study Program at Catholic University. And we participated in this activity. More information in Human Practices/Community |
| | + | Outreach. |
| | + | |
| | + | ==August 27 – September 3, 2012== |
| | + | |
| | + | We transformed pBAD_ADF-3 in pSB1K3, and pBAD_ADF-3+ in pSB1K3. |
| | + | |
| | + | The next day we did colony PCR of those transformed bacteria at a Tm=59ºC. From all the colonies that we used, only one of pBAD_ADF-3 in pSB1K3 and one of pBAD_ADF-3+ in pSB1K3 had the correct size. Then we left these bacteria growing, so that we could purificate their DNA later . |

Contents
- 1 March, 5 - 11, 2012
- 2 March, 12 - 18, 2012
- 3 March, 19 - 25, 2012
- 4 March 26 - April 11, 2012
- 5 April 2-8, 2012
- 6 April 16-22, 2012
- 7 April 23-29, 2012
- 8 April 30 – May 6, 2012
- 9 May 7 – 13, 2012
- 10 May 14 – 20, 2012
- 11 May 21 – 27, 2012
- 12 May 28 – June 3, 2012
- 13 June 4 – 10, 2012
- 14 June 11 – 17, 2012
- 15 June 18 – 24, 2012
- 16 June 25 – July 1, 2012
- 17 July 2 – 8, 2012
- 18 July 9 – 15, 2012
- 19 July 16 – 22, 2012
- 20 July 23 – 29, 2012
- 21 July 30 – August 5, 2012
- 22 August 6 – 12, 2012
- 23 August 13 – 19, 2012
- 24 August 20 – 26, 2012
- 25 August 27 – September 3, 2012
|
March, 5 - 11, 2012
The whole team met for the first time after the vacations of February. We discussed our scheduling and divided ourselves in two groups: Cyanolux and Bactomythril. The leaders of each group were elected: Carla (Cyanolux) and Claudia (Bactomithril).
Cyanolux: Wetlab: Simon, Seba / Drylab: Carla, Tamara, Isaac
Bactomithril: Wetlab: Ulises, Bryon / Drylab: Claudia, Emilia, Max
Carla and Max were chosen to maintain the wiki updated.
Bernardo and Rolando are going to collaborate with both groups.
Also, we scheduled meetings of the whole team every Friday or Tuesday (depending on the availability of the majority of the team), and meetings with the whole team and our advisor, Professor Rodrigo Gutierrez, every other Thursday. Likewise, each group should have their own meetings.
Finally, we decided to work based on delivery dates. Each member should compromise to complete a task in specific time intervals.
March, 12 - 18, 2012
We registered our team in iGEM. Now we are officially part of iGEM 2012! Also we began to learn how to use our wiki.
Our Bactomithril project is still in its initial stage, so we began to read as many papers about recombinant spider silk and protein modeling as we could.
Some of the most relevant papers we found are
- Xia et al. 2010 - “Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber”
- Widmaier et al. - 2009 - “Engineering the Salmonella type III secretion system to export spider silk monomers”
We learnt about interesting protein modeling and simulation software (e.g. Rosetta, Foldit), and some protein visualization software (PyMOL, VMD, Chimera)
Additionally, we began to send emails requesting genetic sequences from spider silk to scientists from other countries. According to what we know, in Chile nobody works with natural or recombinant spider silk proteins or its genetic sequences. So that this is a real challenge to us.
March, 19 - 25, 2012
The iGEM team Slovenia 2009 had a project potentially useful for our own project. For that reason, we sent an email to them asking if they could share their biobricks Bba_K245005 and Bba_K245113 with us.
They answered us and offered their help. But their shipment never arrived. Anyway, later we realized that those parts are not fully suitable for our needs.
Also, some of us had the privilege to assist to a lecture of Andrew Hessel, an authority in Synthetic Biology from USA. Unexpectedly, he had a slide about us! (He found out about us in Internet). He said he was surprised that Chile had an iGEM team working in a forefront discipline such as Synthetic Biology.
But most importantly, we designed our first secretion constructs, consisting of a GFP Reporter Plasmid, a Protease Producer Plasmid (see section constructs for more information) and a working plasmid. This last plasmid is used as an intermediate plasmid to build the other two plasmids. We ordered the primers that we need (it takes about two weeks until they arrive!)
March 26 - April 11, 2012
We digested with restriction enzymes the HIV cleavage site, and transformed an purified the following parts of the Distribution kit 2011:
I712015 HIV 1 cleavage site
I712667 HIV aspartyl protease
I746908 sfGFP
K103006 OmpA
We met with Ignacio Ibarra, an undergraduate student of the group of Dr. Francisco Melo. He is willing to advise us in the area of protein modeling. The problem is that spider silk proteins are semi-crystalline and is difficult to predict its structure. He recommended starting with a comparative protein analysis between our spider silk monomer of interest and other similar proteins that have its structure available in PDB. The idea is that from the structure of those similar proteins we will have a reference of the most stable conformation of our monomer. Later we should be able to improve the self-assembling of the proteins or improve some mechanical properties, such as elasticity and tenacity. Nevertheless, we were warned: protein modeling is a complex and slow task.
Recommended software:
-MOE: to add hydrogen atoms.
-Autodock: a suite of automated docking tools. This should be useful to evaluate the self-assembly of our monomers.
-Force Field: to calculate potentials between particles.
Finally, something very important happened: Christopher A. Voigt, author of “Engineering the Salmonella type III secretion system to export spider silk monomers” is going to help us with a valuable gift: a plasmid to secrete ADF-3 in salmonella. With this plasmid we can obtain the sequence of the spider silk protein ADF-3.
April 2-8, 2012
We transformed the backbone pSB1K3, so that we could amplify it when our primers arrive. Later we made and ordered the primers for our next experiment: basically the same constructs as before (GFP Reporter Plasmid) but with ADF-3 instead of sfGFP.
Ulises met with Dr. Angélica Fierro, and she accepted to be our advisor in protein modeling!
Besides, we contacted Cheryl Hayashi, a well-known scientist that works with spider silk (she even spoke at TED!) [link: http://www.ted.com/talks/cheryl_hayashi_the_magnificence_of_spider_silk.html]
We asked her if she could send us the genetic sequences EF595245 and EF595246, but later we knew that those parts are not compatible with our purposes. They are fosmids and the silk sequence is only a small portion of those clones. The fosmids are low copy number and difficult to propagate. Then, it´s better to work with ADF-3.
Additionally, we wrote to Dr. Sang Yup Lee, author of one of the paper “Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber”. They produced and spun into fiber a 284.9 kDa recombinant protein of the spider Nephila clavipes in E. Coli, and we asked if he could send us his spider silk protein. Unfortunately, we didn´t receive an answer.
April 9-15, 2012
The School of Engineering at Pontificia Universidad Católica de Chile makes every year a fair of entrepreneurship and technological innovation called Ingenia. The deanship of the Faculty of Engineering is so interested in us that wants that we form part in a conversation table to spread information about iGEM and Synthetic Biology. We will have also our own stand in the fair!
On Wednesday the whole team had a meeting with Professor Rodrigo Guitérrez, and our advisors Dr. Fernán Federici (Postdoc student at Cambridge) and Dr. Mónica Vásquez (specialist in cyanobacteria). We presented them our projects and advances.
They suggested that we:
Must not forget the human practices
Should characterize existing biobricks
Should delimit and focus our projects: the time and resources are limited.
Have to consider that protein modeling is a complex task that usually takes a long time to develop.
After the meeting we decided to change our responsibilities.
Wetlab: Bryon and Max
Modeling: Emilia and Ulises
Wiki: Claudia
And finally our primers arrived!! (after 3 weeks). We did PCR of the different parts that we need to build our first construct (the Working Plasmid). For that we:
a) Obtained the parts necessary to make the Working Plasmid and transformed e. coli with them.
b) Did minipreps of all the constructs to purify the DNA from the transformed bacteria.
c) Amplified the constructs in the thermocycler.
d) Did electrophoresis of the PCR amplicons.
We had several problems with the electrophoresis, but after some attempts it worked.
5 of the 7 PCR were successfully amplified. The two parts that didn´t work are HIV cleavage site (2kb) and vector backbone pSB1K3 (42kb).
April 16-22, 2012
We did electrophoresis with HIV cleavage site and pSB1K3. This time, we added more of them (4 μL template in PCR) to ensure that they were present in our tubes. We had now all the parts that we need to proceed!
We made our first Gibson Assembly to build the working plasmid (see constructs). We need this plasmid to build the GFP Reporter Plasmid and the Protease Producer Plasmid. But unluckily our colonies resulted to be red. This is not what we expected… later we did an analytical digestion to the DNA of those colonies, but we obtained strange results. Something went wrong.
This week we had the great opportunity to assist to a lecture of Dr. Aldo Leal. He is a Chilean biochemist that currently works in Germany studying the interactions between cells and biomaterials made of spider silk proteins. We learnt a lot of information about the production of recombinant spider silk proteins and the possibility to easily create films. Then, it’s not necessary to utilize other sophisticated spun methods like electrospinning.
Later we met with Dr. Leal, and he recommended us the following very important paper: Cooper et al. 2009 – “A protocol for the production of recombinant spider silk-like proteins for artificial fiber spinning”.
April 23-29, 2012
We did a new Gibson Assembly to build our working plasmid. This time it worked! We obtained some twenty or thirty colonies.
Additionally, we made our first video with 3DS Max, and Max almost destroyed his notebook with that amount of processing! You can see it here http://www.youtube.com/watch?v=Je5JkoOMITE. Hopefully we will use these tools to make our wiki more attractive.
Also, this week we participated in Ingenia Fair. You can find more information in our wiki section Human practices/Ingenia Fair.
April 30 – May 6, 2012
Analytical PCR was done to verify if the parts that we obtained from the Distribution Kit 2011 to build the GFP Reporter and Protease Producer plasmids have they corresponding size. The results were positive. Then we proceeded with the PCR products purification. Finally we did the Gibson Assemblies to build the GFP Reporter Plasmid and the Protease Producer Plasmid, using pSB1K5 and sfGFP as positive controls. The Assemblies worked all right!
Afterwards, we sent all our constructs (Working Plasmid, GFP Reporter Plasmid and Protease Producer Plasmid) for sequencing.
Also, we are very happy because we received the free license of Matlab. Hopefully we will use this tool for modeling.
May 7 – 13, 2012
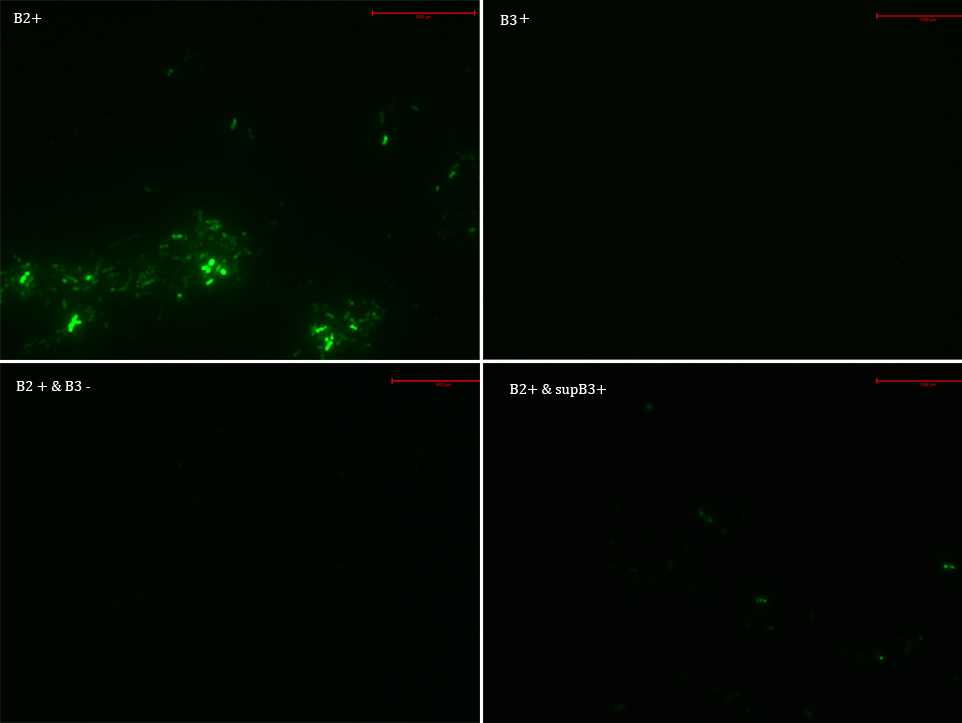
We transformed E. Coli with our GFP Reporter Plasmid (B2) and with the Protease Producer Plasmid (B3). We did a little experiment with those bacteria.
The following samples were prepared:
B2
B2+
B2+ & B3
B2+ & supB3
Nomenclature
B2- (coli transformed with GFP Reporter Plasmid)
B2+ (coli transformed with GFP Reporter Plasmid, induced with Arabinose)
B3+ (coli transformed with Protease Producer Plasmid, induced with Arabinose)
supB3+ (supernatant of coli transformed with Protease Producer Plasmid induced with Arabinose)
Then we observed each sample with an epifluorescence microscope. The results were promising. B2- did not express sfGFP, but B2+ did. Additionally, in both cases (B2+ & B3) and (B2+ & supB3) the fluorescence of sfGFP disappeared almost completely. We are going to do soon another experiment but with a rigorous protocol.
May 14 – 20, 2012
The Distribution Kit 2012 arrived! Now we have a lot of new parts!
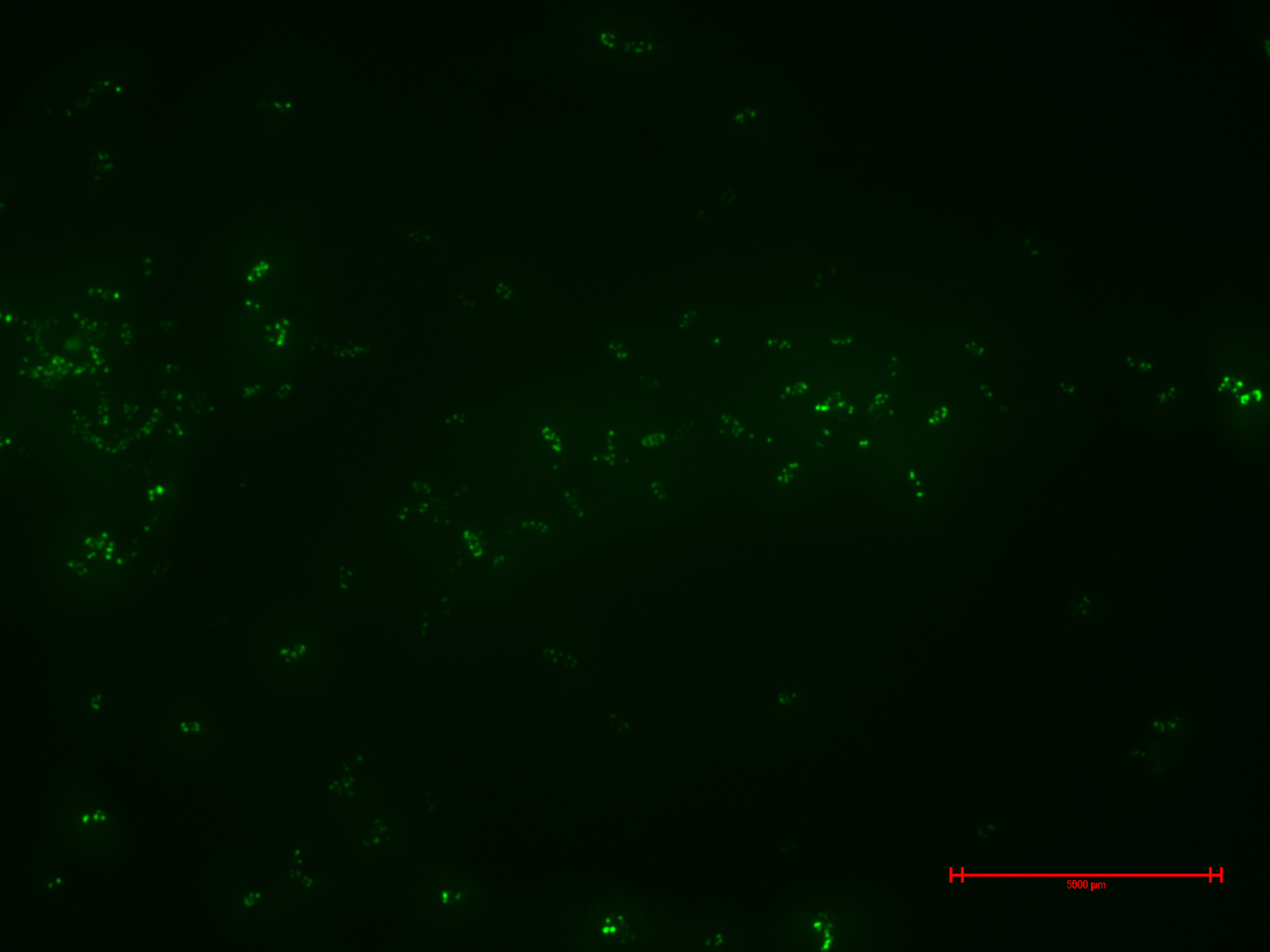
We prepared for second time samples of B2+ & B3+ to observe in the epifluorescence microscope the effect of time in the samples.
This time B2+ & B3+ presented much more fluorescence, and it persisted over time (at least 1 hour with no significant variation).
Additionally, we began to amplify the spider silk monomer ADF-3 from the plasmid that Christopher Voigt gave to us. But the amplicon was barely visible in the electrophoresis. We will have to try it again, or maybe try another technic like touch-down PCR. Or the primers are designed for repetitive areas?
May 21 – 27, 2012
We received the results of the DNA sequencing of Working Plasmid, GFP Reporter Plasmid and Protease Producer Plasmid. Fortunately, these results are coherent with our designs. Great news!
Also, we went to the confocal microscope
May 28 – June 3, 2012
For second time we tried to amplify the spider silk monomer ADF-3 from the plasmid that Christopher Voigt gave to us. This time we believe that the PCR worked!
June 4 – 10, 2012
On Tuesday Bernardo presented all the advances of iGEM team UC Chile 2012 to professor Rodrigo Gutierrez and the Plant Systems Biology Lab members. We received many useful recommendations.
June 11 – 17, 2012
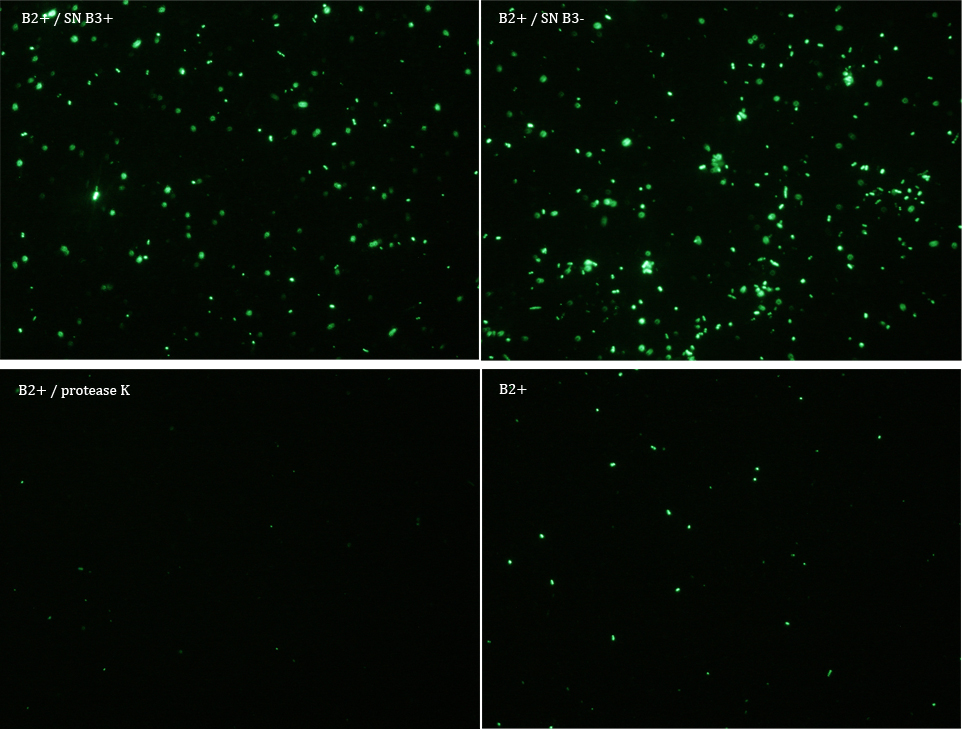
We did another experiment to investigate the interactions between our bacteria B2+ and B3, but this time with a serious protocol and recording the data for later analysis.
We expected that our results were coherent with our theory. [1]. Protease K should also be able to cut the HIV cleavage site, releasing the sfGFP.
The samples used this time were:
B2+ & supernatant B3+
B2+ & supernatant B3-
B2+ & protease K
B2+
Nomenclature
+ induced with Arabinose
- not induced with Arabinose
B2 (coli transformed with GFP Reporter Plasmid)
B3 (coli transformed with Protease Producer Plasmid)
SN supernatant
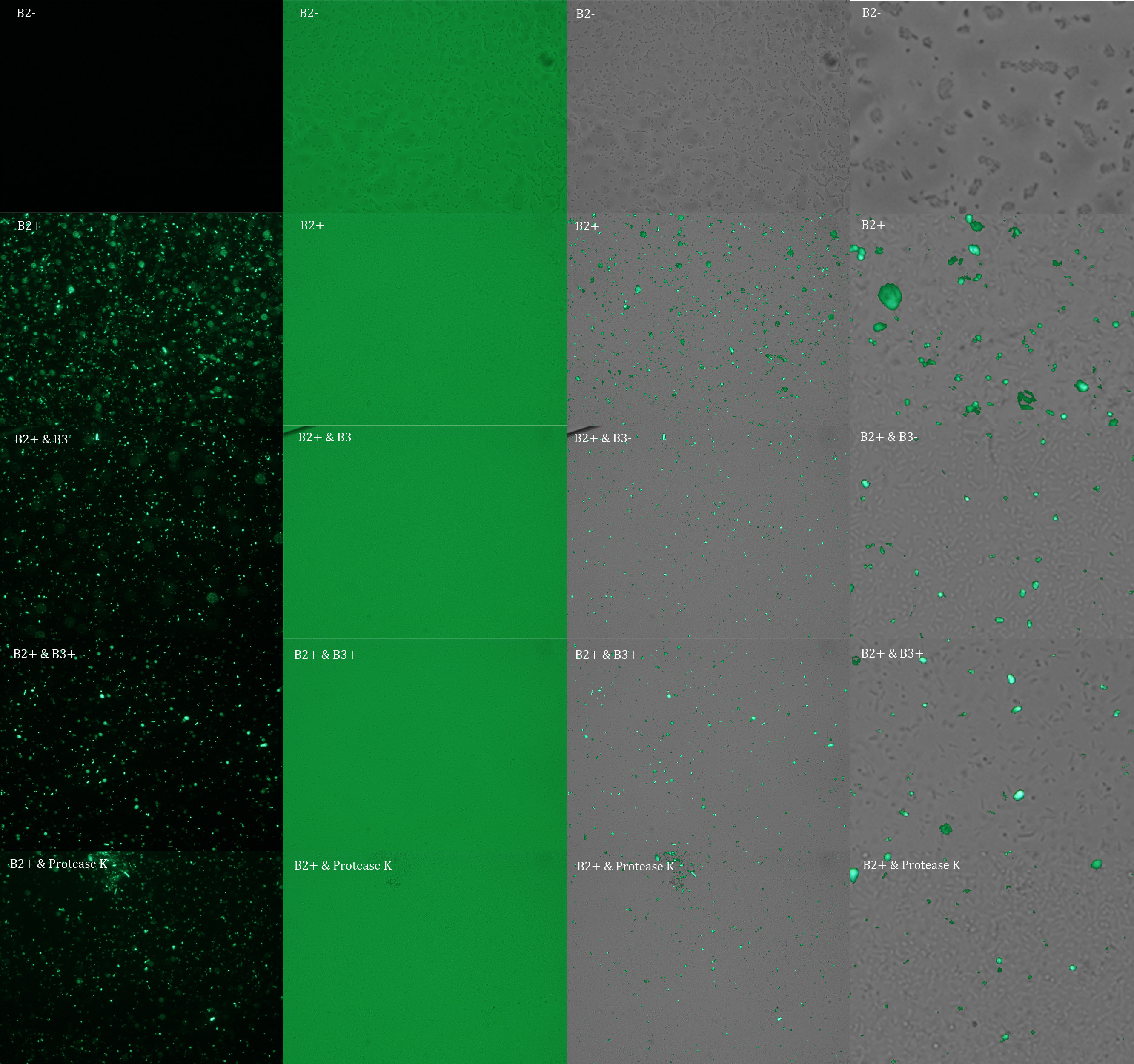
Subsequently we proceeded to observe the samples in an epifluorescence microscope. In the following figure you can se one micrograph of each sample at 100x.
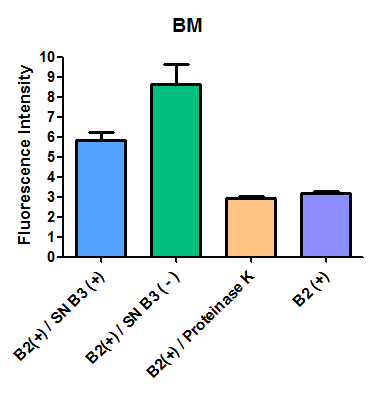
Additionally, we analyzed 4 micrographs of each sample (n=4) with the software ImageJ
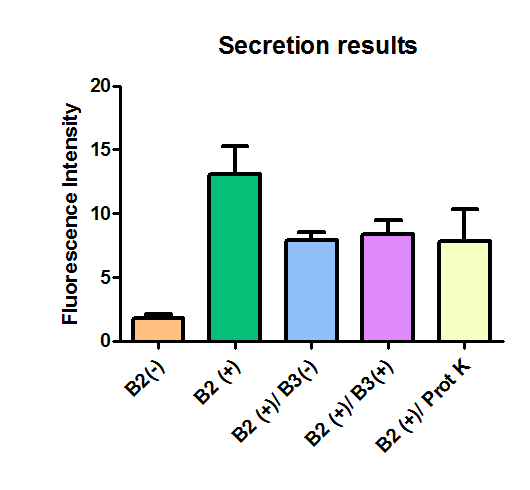
All the different treatments presented significant differences, except (B2+ & protease K) and (B2+). Remarkable is the difference between (B2+ / SN B3+) and (B2+ / NS B3-). Also, the direct application of protease K over the samples decreased even more the fluorescence. However, the low fluorescence of B2+ is very weird. We will have to repeat this experiment, but at least the major part of the results makes is what we expected.
June 18 – 24, 2012
We repeated the experiment to investigate the interactions between our bacteria B2+ and B3+ with a serious protocol and recording the data for later analysis. This time we added the control B2-.
In the following figure you can se one micrograph of each sample at 100x.
In the figure each row represents one sample. The first row contains the micragraphs with UV filter. The second row contains the micrographs seen with light. The third row contains the previous micragraphs overlayed (the light image is in grayscale, and the UV image has its black background removed). The forth image is an amplification of the third image (1:4 ratio). Click over the image to open it in a higher resolution.
Additionally, we analyzed 4 micrographs of each sample (n=4) with the software ImageJ
The results were quite different compared to the ones of our previous experiment. Remarkable is that, as expected, B2- and B2+ present the lowest and highest fluorescence intensity. Additionally, (B2+ & B3+) and (B2+ & Protease K) present very similar fluorescence. However, for second time we obtained a weird result: (B2+ & B3+) and (B2+ & B3-) present the same amount of fluorescence.
In conclusion, we know that sfGFP expresses in our e. coli. But we don’t know if it is attached to the external side of the membrane, or if the HIV-1 protease works as expected. As this secretion system seems to be failing, we chose to try other alternatives.
June 25 – July 1, 2012
The Gibson Assemblies of our iGEM team had been failing in the last weeks. This week new enzimes arrived. We hope that this will solve our problem.
Also, this week we tried to redesign our project. Our first task is to continue with our attempts to make a biobrick with the spider silk monomer ADF-3. We ordered the primers that we need to do that.
Now we are exploring the possibility of improve the product of the secretion system developed by Christopher Voig in his paper “Engineering the Salmonella type III secretion system to export spider silk monomers”. One idea is to attach a growth factor (e.g. recombinant human BMP-2) to the molecule of ADF-3.
But the problem with the secretion system developed by Christopher Voig is that it works on salmonella, and we cannot work with such an infectious bacteria.
July 2 – 8, 2012
This was a week of exams. Therefore, no major advances were accomplished during this week.
July 9 – 15, 2012
This week we made important advances in our wiki. We will start with the template of the iGEM team Johns Hopkins 2011 as a base.
Furthermore, we studied carefully the paper “Engineering the Salmonella type III secretion system to export spider silk monomers” and its supplementary information. We realized that each protein secreted with this system comes with a 3x FLAG (DYKDDDDK)3. It is a priority for us to find if someone can share us a little of the anti-FLAG antibody (Sigma Cat# F3165). That way, if we use this secretion system we will be able to quantify the production of our protein of interest.
But, if we want to use Voigt´s system, we need attenuated salmonellas with their pathogenicity island 1 intact.
Something similar to Salmonella Typhimurium SL1344. After some research we found a professor of our University that works with salmonella (Dr. Susan Bueno). But she was out of the country and we could only send her some emails asking for the salmonella.
July 16 – 22, 2012
Finally the primers necessary for biobricking ADF-3 arrived!
And we have got other important material: we talked with Dr. Hugo Olguin, and he is willing to share with us his anti-FLAG antibody. Furthermore, he allowed us to do the western blot in his laboratory.
Additionally, we achieved significant advances in the wiki, especially in the section Human Practices.
Unfortunately, we still lack of salmonella.
July 23 – 29, 2012
After some research we found the promising projects of the Japanese iGEM team HokkaidoU 2010 and 2011. They used the E. Coli (k-12) strain SGSC4024, from Salmonella Genetic Stock Center(SGSC) in University of Calgary, Canada (http://people.ucalgary.ca/~kesander/index.html). This strain carries a pBeloBAC11 vector encoding a genome fragment of Salmonella enterica serovar Typhimurium LT2 which covers the SPI-2 region [B_STM07H21 SGSC4024 1464540~1562427]
We asked the iGEM team members of HokkaidoU 2011 if the E. Coli (k-12) strain SGSC4024 is capable of secreting proteins to the external environment. But unfortunately it seems that is only capable of injecting molecules into other cells, and that is not what we need.
In the lab we did PCR to amplify ADF-3 and ADF-3+ (i.e. ADF-3 with the tags included in the plasmid that Christopher Voigt sent to us). Unfortunately the PCR did not work.
The next day we tried to amplify the backbone pSB1C3, ADF-3+ and ADF-3 with touch-down PCR. The two first amplicons seemed to be all right. Nevertheless, later we purificated and digested the DNA to verificate if the amplicons corresponded to our parts, but the size sequences resulted to be incoherent. We have to try again.
July 30 – August 5, 2012
This week we did PCR of the backbone pSB1C3, ADF-3+ and ADF-3 again. It failed again. Are our primers wrong?
However, we persisted tirelessly with another PCR of pSB1C3, ADF-3+ and ADF-3, this time with a Tm=59º C. It worked for ADF-3+!!
We purified the DNA sequence of ADF-3+. Afterwards we did PCR again, but this time to amplify the backbone of ADF-3 that has the promotor pBAD, and to amplify the vector backbone for ADF-3+.
The PCR of the backbone of ADF-3 that has the promotor pBAD looks successful! Later we proceeded with the corresponding DNA purification.
Joyfully we proceeded with the Gibson Assembly to join ADF-3 to its backbone with pBAD. We transformed the E. coli, but no colony survived.
August 6 – 12, 2012
We did another PCR, but this time amplified ADF-3 with a Tm=56º C ; The vector backbones of ADF-3+ and ADF-3 with Tm=60º C and touch-down PCR of the vector backbones of ADF-3+ and ADF-3 with T annealing = 54º C.
Tragically, none of them worked.
The next day we did PCR again with Tm=59ºC of ADF-3 and the vector backbones of ADF-3+ and ADF-3.
The PCR of ADF-3 was successful! Later we did the purification of the DNA.
Afterwards, we completed the Gibson Assambly of ADF3 with the backbone PSB1C3 and the transformations of the E. coli.
The next day finally we had some transformed E. coli! Immediately we proceeded with colony PCR to verify if the transformed bacteria correspond to our designs. Unluckily, the colony PCR was negative (the DNA strand that we obtained had a smaller molecular weight than what we expected).
So that we repeated the PCR for the vector backbones of ADF-3 and ADF-3+. The bands were from the right length! Afterwards we proceeded with the Gibson Assembly of ADF-3 with its backbone, and of ADF-3+ with its backbone. Both Gibson Assemblies were positive! Finally we continued with a colony PCR.
August 13 – 19, 2012
We purificated the DNA from the colony PCRs that were successful, and then digested it. Fortunately one digestion of ADF-3 and other of ADF-3 had the correct size.
Later we did PCR to amplify the backbones of ADF-3 and ADF-3+ (both with pBAD). But we made some errors and added the wrong primers.
The next day we repeated the PCR, and the backbone with pBAD of ADF-3 had the right size.
Then we send for sequencing ADF-3 and ADF-3+. Later we realized that we sent the wrong sample of ADF-3!!
We also transformed some E. coli with backbone with pBAD of ADF-3. The following day we did colony PCR to these E. coli, but we did not obtain the desired results. Then we cultivated the E. coli transformed with the backbone with pBAD of ADF-3 in 200 mL of LB with 1nM of arabinose.
August 20 – 26, 2012
We did colony PCR of 3 colonies transformed with the backbone with pBAD of ADF-3, but the results were negative.
The next day we did colony PCR again, but this time with other method:
1) Put 100 mL of bacteria in eppendorf tubes of 2µl and heat at 95ºC for 10 minutes.
2) Centrifuge at maximum speed for 5 minutes.
3) Do Colony PCR with the supernatant as the template.
Unfortunately, the results were negative.
And we became the results from the sequencing. ADF-3 is incorrect (as we expected, because we sent the wrong sequence), and ADF-3+ is a bit weird. Bernardo never trusted the PCR band of this sequence anyway…
The next day we sent for sequencing ADF-3 (this time the right sequence). Some days later we became the results from this sequencing, and it were negative. Dammit!
Finally, we prepared the materials for the community outreach activity for Penta UC, the Talents Development and Study Program at Catholic University. And we participated in this activity. More information in Human Practices/Community
Outreach.
August 27 – September 3, 2012
We transformed pBAD_ADF-3 in pSB1K3, and pBAD_ADF-3+ in pSB1K3.
The next day we did colony PCR of those transformed bacteria at a Tm=59ºC. From all the colonies that we used, only one of pBAD_ADF-3 in pSB1K3 and one of pBAD_ADF-3+ in pSB1K3 had the correct size. Then we left these bacteria growing, so that we could purificate their DNA later .
 "
"