Team:Cambridge/Ratiometrica/Labbook
From 2012.igem.org
Contents |
Judging Form
- Please help the judges by filling out this form. Tell them what medal you think you deserve and why. Tell them which special prizes you should win. Help them find your best parts. Show them how you thought about the safety of your project. Helping the judges will help you too.
- Team: Cambridge
- Region: Europe
- iGEM Year:2012
- Track:Foundational Advance
- Project Name:Parts for a reliable and field ready biosensing platform
- Project Abstract: Implementation of biosensors in real world situations has been made difficult by the unpredictable and non-quantified outputs of existing solutions, as well as a lack of appropriate storage, distribution and utilization systems. This leaves a large gap between a simple, functional sensing mechanism and a fully realised product that can be used in the field.
We aim to bridge this gap at all points by developing a standardised ratiometric luciferase output in a Bacillus chassis. This output can be linked up with prototyped instrumentation and software for obtaining reliable quantified results. Additionally, we have reduced the specialized requirements for the storage and distribution of our bacteria by using Bacillus' sporulation system. To improve the performance of our biosensing platform we have genetically modified Bacillus’ germination speed. Lastly, we demonstrated the robustness of our system by testing it with a new fluoride riboswitch, providing the opportunity to tackle real life problems.
iGEM Medals for non-software teams
- We believe our team deserves the following medal:
- Bronze
- Silver
- √Gold
Because we met the following criteria (check all that apply and provide details where needed)
Requirements for a Bronze Medal
- √Register the team, have a great summer, and plan to have fun at the Regional Jamboree.
- √Successfully complete and submit this iGEM 2012 Judging form.
- √Create and share a Description of the team's project using the iGEM wiki and the team's parts using the Registry of Standard Biological Parts.
- √Plan to present a Poster and Talk at the iGEM Jamboree.
- √Enter information detailing at least one new standard BioBrick Part or Device in the Registry of Standard Biological Parts. Including:
- √Primary nucleaic acid sequence
- √Description of function
- √Authorship
- Safety notes, if relevant.
- √Acknowedgment of sources and references
- √Submit DNA for at least one new BioBrick Part or Device to the Registry.
Additional Requirements for a Silver Medal
- √Demonstrate that at least one new BioBrick Part or Device of your own design and construction works as expected; characterize the operation of your new part/device.
- √Enter this information and other documentation on the part's 'Main Page' section of the Registry
Part Number(s): BBa_K911004
Additional Requirements for a Gold Medal: (one OR more)
- Improve an existing BioBrick Part or Device and enter this information back on the Experience Page of the Registry.
Part Number(s): None - √Help another iGEM team by, for example, characterizing a part, debugging a construct, or modeling or simulating their system.
Link to this information on your wiki. Page name: Team:Cambridge/Outreach/Collaboration - √Outline and detail a new approach to an issue of Human Practice in synthetic biology as it relates to your project, such as safety, security, ethics, or ownership, sharing, and innovation.
Link to this information on your wiki.
Page name: Team:Cambridge/HumanPractices/Overview,Team:Cambridge/HumanPractices/MarketResearch,Team:Cambridge/HumanPractices/FutureDirections
iGEM Prizes
All teams are eligible for special prizes at the Jamborees. more... To help the judges, please indicate if you feel you should be evaluated for any of the following special prizes:
- √Best Human Practice Advance
- √Best Experimental Measurement
- Best Model
Please explain briefly why you should receive any of these special prizes:
Best Human Practice Advance:
We feel that we deserve this prize for three reasons:
- We explored the impacts, *both positive and negative*, of synthetic biology as a solution to real world problems, through interviewing professionals working in a relevant field, namely the impact of arsenic water contamination in Bangladesh.
- We recognized existing problems with the way the current direction of synthetic. On going through the registry we found that most of the characterization data for biosensing parts is often neither comparable nor replicable. We have worked to solve this issue, for example with our ratiometric dual channel output.
- *Our project doesn’t stop here*, in Chanel number 6 (Team:Cambridge/HumanPractices/FutureDirections) we considered the future implications and technological applications of our project, as well as the means by which it could be improved by subsequent users. We feel that the end to an iGEM project should not be the conclusion of an idea, but the start of it.
Best BioBrick Measurement Approach:
It is absolutely vital that a quantitative, numerical, robust, and flexible measurement approach exists to relay information to a user that is an accurate representation of the input processed by a biological device. Working from these principles, the following was done:
- We designed and built Biologger, a *cheap, arduino-based, fully functional automatic rotary device* that has an incorporated ratiolumnometer
- Our project is entirely open-sourced and open-platform. We have published source code for the two applications which serve to operate the device, one for PCs and the other for Android devices, as well as the open source circuit design that provides this ratiometric reading. Furthermore, the Android app is able to receive its data wirelessly, which we feel is a great advance in BioBrick measurement.
- Our dual-channel luciferase reporter was successfully tested with a dilution series of E.coli transformed with the Lux Operon (under pBAD) biobrick (Part BBa_K325909) of the Cambridge iGEM 2010 team. It can detect, with good accuracy, both different light intensities, as well as the percentages of blue or orange frequencies in a sample.
- Our device was successfully tested using artificial light to detect different frequencies (colours) as well.
Having done all the above, we believe that this fully open-sourced instrumentation kit (mechanical) chassis, electronics, software code), estimated at *$35.00* (or $85.00 if a Bluetooth modem is required), is a complete BioBrick measurement solution for any and all BioBricks with a light output.
Team_Parts
To help the judges evaluate your parts, please identify 3 of your parts that you feel are best documented and are of the highest quality.
- Best new BioBrick part (natural)
- BBa_K911003
- Best new BioBrick part (engineered)
- BBa_K911004
- Best improved part(s): None
List any other parts you would like the judges to examine:BBa_K911001, BBa_K911009, BBa_K911008
Please explain briefly why the judges should examine these other parts:
- Magnesium Sensitive Riboswitch BBa_K911001
As a riboswitch sensing construct, this part is an entirely new type of biosensor (along with the fluoride construct) that could potentially change the way we think about designing input genetic circuits. Unlike the fluoride riboswitch, it is a derepression system and therefore serves to demonstrate the principle that riboswitches can be used regardless of whether they turn on or off their reporter. - Fluorescent ratiometric construct for standardizing promoter output BBa_K911009
Fluorescence is a major cornerstone for biosensors in the registry, however, most parts do not involve the use of a ratiometric output, which has been shown in the literature to provide much more reliable and meaningful data. This part not only furthers the development of ratiometric measurements in molecular biology but due to the choice of promoters and terminators it can be used to characterize the difference in activity between E. coli and B. Subtilis - Fast Germination (B. subtilis) BBa_K911008
This part is entirely novel for the registry and fully utilizes the recombination machinery inherent in the Bacillus chassis. Have spores that can germinate at a faster rate is certainly a worthy achievement and could help with experiments with B. Subtilis that any future iGEM teams may wish to perform.
iGEM Safety
For iGEM 2012 teams are asked to detail how they approached any issues of biological safety associated with their projects.
The iGEM judges expect that you have answered the four safety questions Safety page on your iGEM 2012 wiki.
Please provide the link to that page: Page name: Team:Cambridge/Safety
Attribution and Contributions
For iGEM 2012 the description of each project must clearly attribute work done by the team and distinguish it from work done by others, including the host labs, advisors, and instructors.
Please provide the link to that page, or comments in the box below: Page name: Team:Cambridge/Attributions
Comments
If there is any other information about your project you would like to highlight for the judges, please provide a link to your wiki page here: Team:Cambridge/Overview/DesignProcess
Ratiometrica Labbook
Week 4
Monday (16/07/12)
Ratiometrica- Lux: Transformation of bacillus and e.coli with lux genes from 2010
- Arabinose induction: from Cambridge 2010 iGEM Team's data, the arabinose concentration which will give the optimum amount of light is around 3mM. We hence added 0.45mg arabinose to each mL of LB.
- Light is detected after around 5 hours in the transformed E. coli
- B. subtilis plates have no growth, very possibly due to plasmid backbone pSB1C3 not being compatible in B. subtilis
Wednesday (18/07/12)
Ratiometrica-Lux: Transformation of e.coli with lux genes from 2010
- Progress: (1) Streaked 8 25ug/ml Chloramphenicol plates with transformed e.coli and incubate overnight; (2) inoculate individual colonies from the plates in liquid medium; (3) Incubate overnight at 37 degrees
Friday (20/07/12)
Ratiometrica-Lux: Transformation of e.coli with lux genes from 2010
- Arabinose induction: Using the same 3mM arabinose prepared earlier in the week, the E. coli were spun down and the arabinose was added. After 5 hours, light is observed.
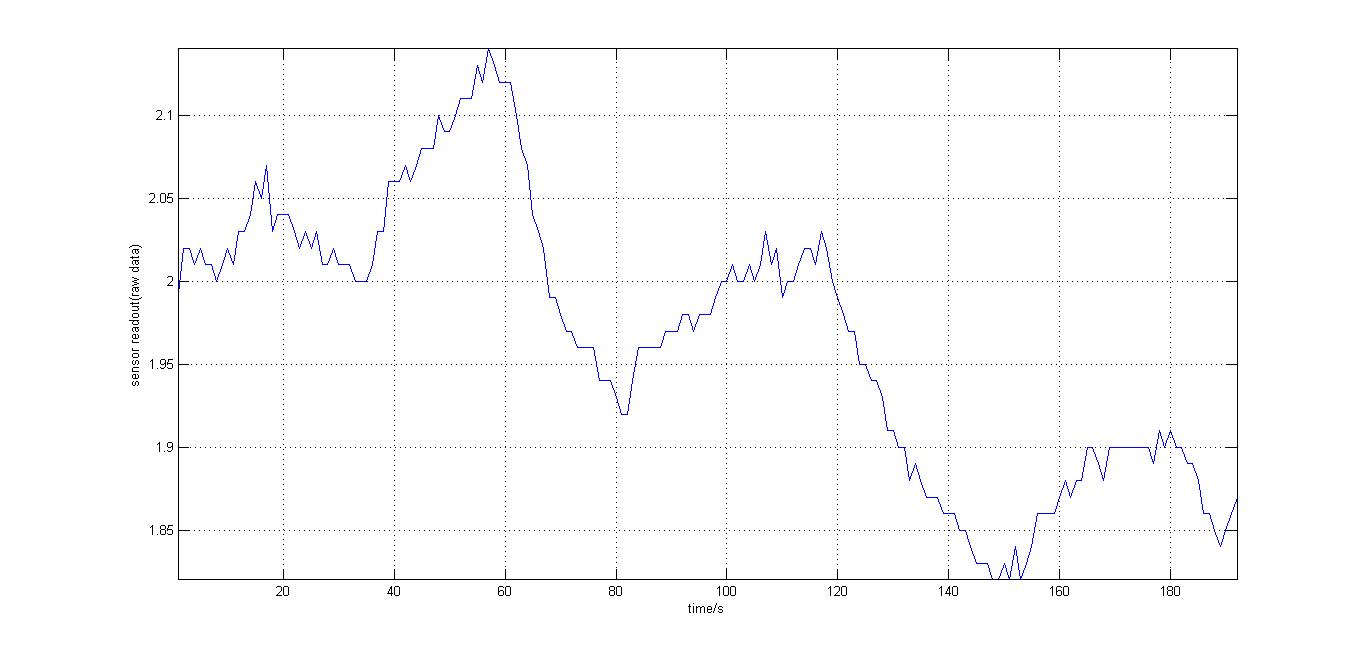
- Testing the Arduino kit: the testing took place in the dark room, and the cell culture (in 1.5mL Eppendorf tubes) was held close to the photosensor and then taken away repeatedly. The data recorded was plotted with Matlab, as shown:
- It should be noted that the background light levels were falling linearly, possibly due to negative feedback on the photosensor, but there is a consistent 5-6% increase of the original base reading upon placing the cells close the the photosensor.
Week 5
Thursday (26/07/12)
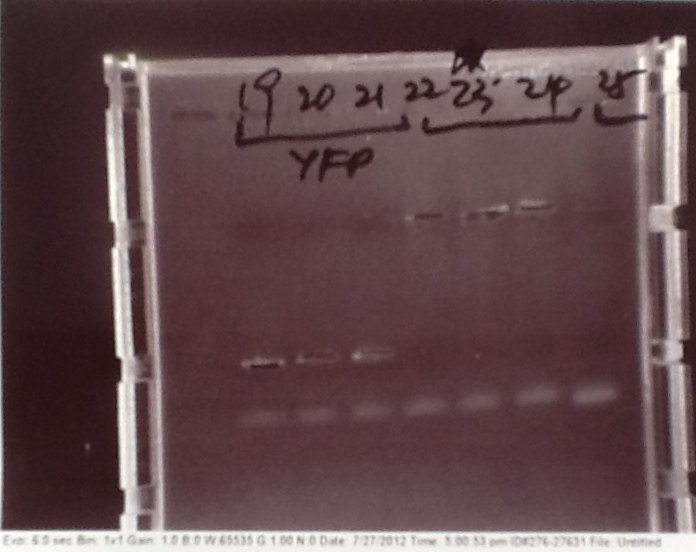
Ratiometrica: PCR of vectors, CFP, YFP, Promotor, Terminator and mOrange
- Plasmid PJS 130 used as our backbone for isolating the magnesium riboswitch and constructing the fluorescent ratiometric plasmid.
- Parts from registry amplified: E0020 (CFP), B0015 (Terminator), K143083 (Pveg) and E0030 (YFP).
- mOrange from lab also used
- Primers:
- E0020
- Forward: cacaattaaaggaggaattcaaa|ATGGTGAGCAAGGGC
- Reverse: tcgttttatttgatgcctgg|TTATTACTTGTACAGCTCGTCCA
- B0015
- Forward: acgagctgtacaagtaataa|CCAGGCATCAAATAAAACGA
- Reverse: aaaattattttgacaaaatt|TATAAACGCAGAAAGGCCC
- K143083
- Forward: tgggcctttctgcgtttata|AATTTTGTCAAAATAATTTTATTG
- Reverse: tcctcgcccttgctcaccat|ctagta|TTCACCACCTTTCTCTAGTAACA
- E0030
- Forward: tgttactagagaaaggtggtgaa|tactag|ATGGTGAGCAAGGGCG
- Reverse: gatgcctggctctagtatca|TTATTACTTGTACAGCTCGTCCA
- mOrange
- Forward: accaaaaaggaatagagt|ATGGTGAGCAAGGGCG
- Reverse: caaacttcat|ACTTCCTCCTCCTCCACTTCCTCCTCCTCC|cttgtaagctcgtccatgc
- pJS130 Mg2+
- Forward: tagaggaggtacgagtcccg|ATGAAACCAGTAACGTTATACGA
- Reverse: tacatcacaattacggaaca|CAAAATCGTCTCCCTCC
- pJS130 Fluorescent
- Forward: acgagctgtacaagtaataa|TGATACTAGAGCCAGGCATC
- Reverse: tcctcgcccttgctcaccat|TTTGAATTCCTCCTTTAATT
- PCR settings:
- 95 °C - 6mins
- 98 °C - 10secs
- 58 °C - 45secs
- 72 °C - 180secs
- Repeat above 35x
- 72 °C - 5mins
- 25 °C - 1min
Friday (27/07/12)
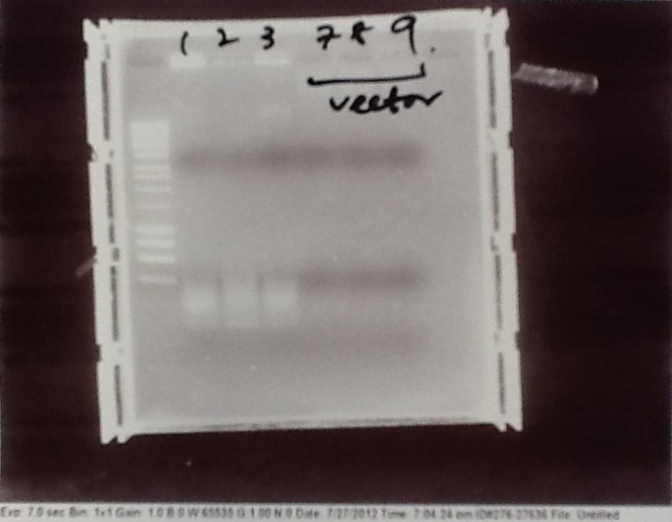
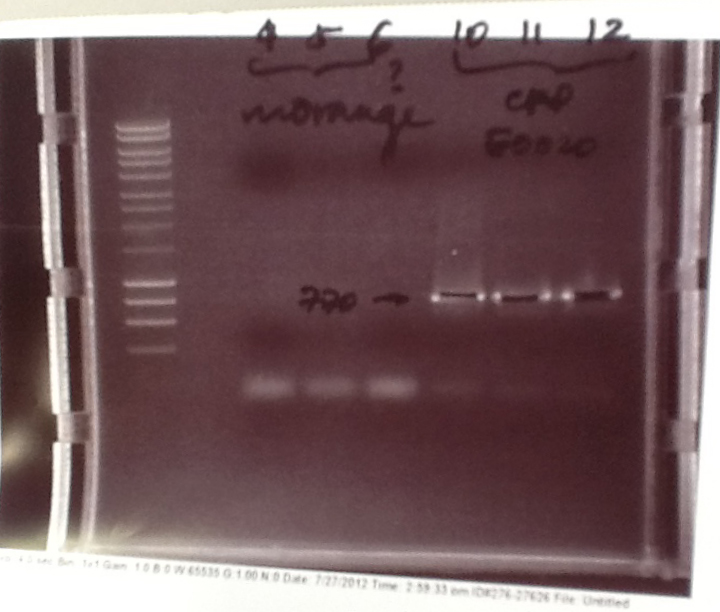
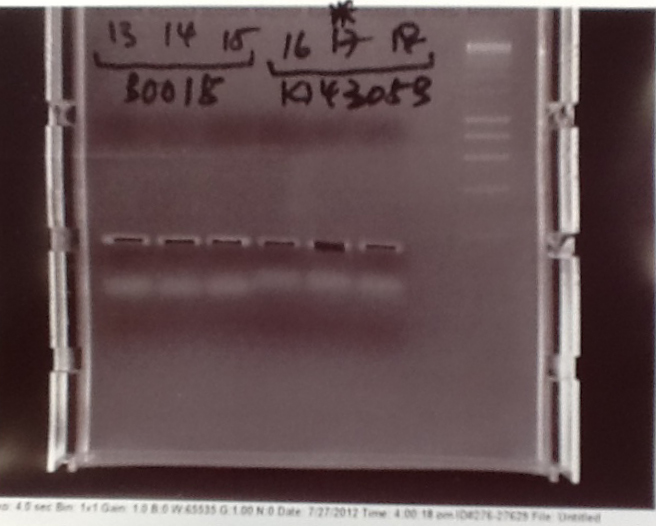
Ribosense & Ratiometrica: Gel electrophoresis of PCR products
- PCR fragments for Mg2+ riboswitch, luxA/mOrange fusion and fluorescent construct from yesterday separated on gels. 90mins at 100V.
- Mostly successful, but will need to repeat four runs: Three vectors (fluorescent, fusion (lux) and Mg2+ riboswitch (-8 codons) and mOrange gene.
Ribosense & Ratiometrica: Purification of successfully amplified DNA from gel
- Successful CFP, YFP, B0015, Pveg + SpoVG (K143053), riboswitch and vector DNA extracted from gel and purified using a minelute column. DNA frozen for later Gibson assembly.
Week 6
Monday (30/07/12)
Ratiometrica & Ribosense: PCR of vectors and mOrange
- Not all products were obtained during Friday's PCR. Most of these missing products were large vector backbones. They are being run again, with a much longer extension time of 300s. If that fails, primers will be ordered to split the vectors into manageable chunks, and the PCR reattempted when they arrive.
- PCR cycle x35:
- 15s Denaturing at 95 C
- 45s Annealing at 60 C
- 300s Extension at 72 C
- Remaining stray product had a slightly tricky secondary structure at the 3' end. It will be run at a series of annealing temperatures in a PCR machine capable of a temperature gradient.
- mOrange PCR run at many different temperatures, from 62 °C to 76 °C. However this doesn't seem to solve our problem (refer to Gel photos). It seems likely that it is a primer design problem.
Ratiometrica: Separation of vector and mOrange DNA
- Positive control produced no band. No primer smear - primers may not be in mix for some reason.
- Realized correct lux vector template was not added during PCR preparation, consequently no amplification occured. Still appears to be a primer smear.
- Fluorescent vector backbone produced several bands. Appears to be due to mis-priming during PCR. Either changing the primers or raising the annealing temperature should solve this problem, but may mean that we have to do this PCR separately.
- Riboswitch vector also failed. However, the extraction from the previous PCR run may have worked. We will try producing a functional plasmid with this extraction before running this PCR again.
Tuesday (31/07/12)
Ratiometrica-Flu: PCR of split fluorescent construct
- Our present theory is that the vectors that we are trying to amplify is too large for the PCR to effectively take place. Given this, we are using split primers to try to reduce the size of the fragments (kindly provided by P.J.Steiner).
- New primer sequences:
- Forward: tgaagtgttcgacaatataaatgtg|CGAAACGATCCTCATCCTGT
- Reverse: ACAGGATGAGGATCGTTTCG|cacatttatattgtcgaacacttca
- PCR Programme
- After gel electrophorsis, it was found that the PCR had worked perfectly. Given we now have all the fragments for the fluorescent construct, we will try to make it with Gibson ASAP.
Friday (03/08/12)
Ratiometrica-Flu: Construction of fluorescent plasmid with Gibson Assembly
- Isothermal reaction buffer *5 remade with correct composition. Needed to order NAD+, so used some master mix from Haseloff lab.
- Various Gibson tail tagged DNA fragments produced over last few weeks assembled together using Gibson protocol:
- pJS130 plasmid fragment A
- pJS130 plasmid fragment B
- CFP (E0020)
- Terminator (B0015)
- pVEG + RBS (K143053)
- YFP (E0030)
Ratiometrica-Flu: Transformation of TOP10 e.coli with Gibson products
- 3 tubes of TOP10 transformed with Gibson products. Cells plated out on 100μg/ml ampicillin plates.
- No positive control added. If no cells grow, will add positive control to all future experiments.
- Results: no growth (04/08/12)
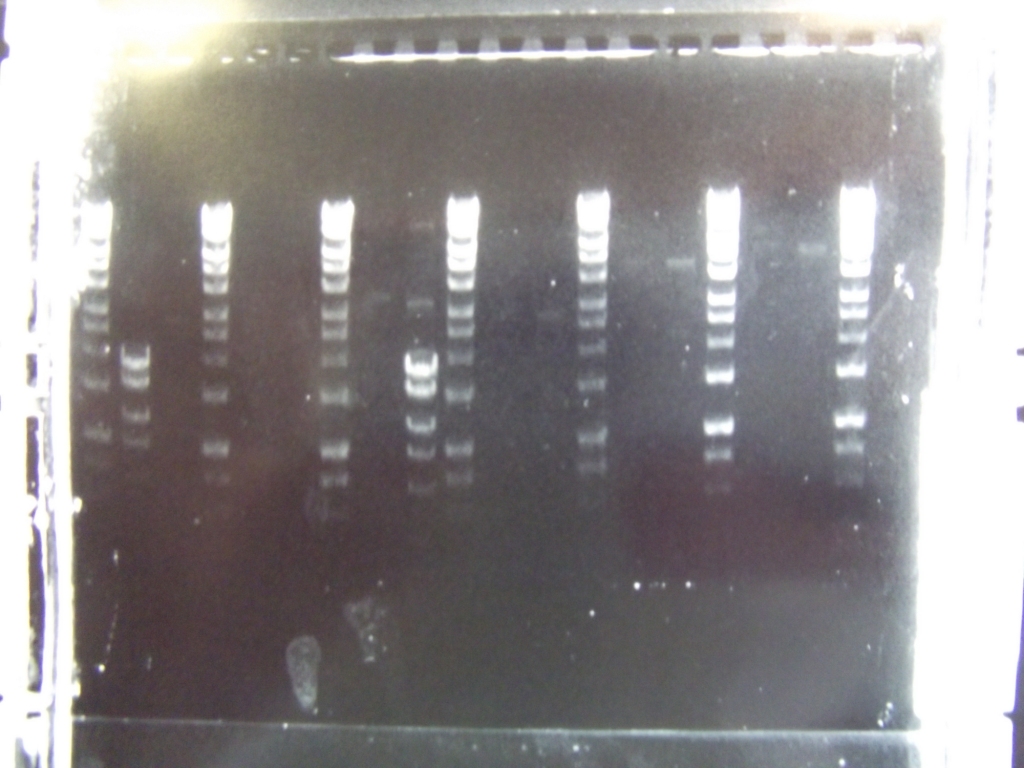
Ribosense: PCR of split Mg2+ vector
- PCR of magnesium riboswitch vector repeated with primers to split plasmid.
- Lanes 2 + 3: Fragment A (center - cut site (promotor side))
- Lanes 4 + 5: Fragment B (without 8 codon substitution) (cut site (lac I side) - center)
- Lanes 6 + 7: Fragment B (with 8 codon substitution) (cut stie (lac I side) - center)
- Gels run, found PCR was unsuccessful.
Ratiometrica-Lux: PCR of mOrange
- with the forward primer correctly labelled and the single-base deletion in the reversed primer fixed, we were able to obtain the mOrange fragment
- PCR Programme:
- Result: bands of the correct size (Lanes 7-8)
Week 7
Monday (06/08/12)
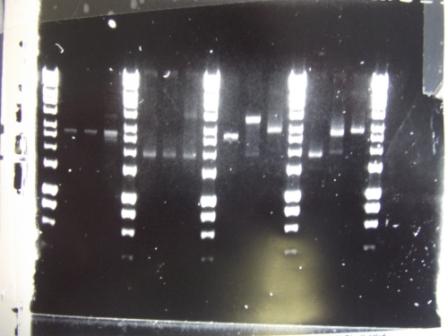
Ratiometrica-Lux: Split PCR of Lux vector
- Primers for spliting pSB1C3 arrived and were used to PCR the Ratiometrica-Lux vector
- Fragment A: Vec FWD and Vec split RVS (expected size: 5kb)
- Fragment b: Vec split FWD and Vec RVS (expected size: 4.6kb)
- PCR Programme:
- Results: other than one very ambiguous band (lane 6) which might be of the right size, the others did not work.
Tuesday (07/08/12)
Ratiometrica-Flu: Gibson assembly of Fluorescent construct
- 6-piece Gibson assembly done in triplicate
- Reaction 1: Tube 1 (Fragment A), Tube 4 (Fragment B), Tube 10 (CFP), Tube 13 (B0015), Tube 16 (K143053), Tube 19 (YFP)
- Reaction 2: Tube 2 (Fragment A), Tube 5 (Fragment B), Tube 11 (CFP), Tube 14 (B0015), Tube 17 (K143053), Tube 20 (YFP)
- Reaction 3: Tube 3 (Fragment A), Tube 6 (Fragment B), Tube 12 (CFP), Tube 15 (B0015), Tube 18 (K143053), Tube 21 (YFP)
Ratiometrica-Lux: colony PCR of Lux vector
- 2nd attempt at amplifying the Lux vector in two fragments, similar to one attempted yesterday
- Results: no bands of the correct size. There seems to be DNA in the well which indicates presence of very large pieces of DNA.
Wednesday (08/08/12)
Ratiometrica & Ribosense: Electrical transformation of competent e.coli with Gibson products
- Gibson constructs from yesterday (fluorescent and magnesium riboswitch) transformed into e.coli produced yesterday.
- Cells plated out onto 50 μg/ml ampicillin plates. Put in incubator overnight.
Ratiometrica-Flu: Restriction Digest of Gibson products
- A restriction digest was performed to investigate if the plasmid was actually successfully constructed in the 6-piece Gibson reaction
- Restrictrion Enzymes: SalI (site at 5619), XhoI (site at 8040)
- Restriction Enzyme buffer: NEBuffer 3
- Results: No bands at all; probably because there was too little DNA.
Week 8
Monday (13/08/12)
Ratiometrica-Lux: Miniprep of LuxBrick plasmid
- Miniprepped is done using miniprep kit supplied by Cambio
- Plasmids are miniprepped from 3 cultures
Ratiometrica-Lux: PCR of lux vector
- With split primers, similar conditions to last week (except no longer using E. coli colonies), each half in triplicates
- Results: Half of the vector (Fragment B) came out (gel 1), positive control worked (gel 2 lane 5)
Ratiometrica-Lux: Arabinose induction of Lux E. coli
- E. coli with the lux plasmid (K325909) O/N liquid cultures are induced with 3mM arabinose
- They did not produce visible bioluminescence after 7 hours. We suspect it to be a problem associated either with the concentration of cells in the culture or the amount of arabinose we added.
Tuesday (14/08/12)
Ratiometrica-Lux: Arabinose Induction of Lux E. coli
- After our failed liquid culture induction, we made LB agar plates with 3mM arabinose and the appropriate antibiotics.
- Cells are plated out on the arabinose plates and incubated at 37°C overnight.
- Colonies on plates show bioluminescence after overnight incubation
Thursday (16/08/12)
Ratiometrica-Lux: Restriction Digest of Lux vector
- Results: Digest came out exactly as expected.
Ratiometrica-Flu: PCR of Flourescent Construct DNA fragments
- Redo of PCR done at the end of July, as we are running out of fragments to Gibson
- Used existing DNA fragments as template rather than biobricks from the registry
- Results: Only the fluorescent protein fragments (eCFP and eYFP) came out clearly; the smaller fragments (B0015, K143053) might have came out but was mistaken for primer dimers, the vector fragments did not come out
Saturday (18/08/12)
Ratiometrica-Flu: 2nd attempt at PCR of fragments
- Reattempt of pJS130 and smaller fragments
- Used Velocity from Bioline, new mastermix recipe is used
- Results: failed- no wanted bands, since the positive control didn't come up, it is suggested that we need to optimise the PCR protocol for Velocity if we are to use it.
Ratiometrica-Flu: Gibson assembly of fluorescent construct
- Gibson performed using remaining DNA fragments which has been concentrated by dehydration
- We realised that though the decreased T5 exonuclease in the gibson mastermix might have lowered the Gibson efficiency in the positive control, the fragments we are trying to assemble are much smaller (~200bp); papers said this is likely to cause trouble
- We therefore attempt 2 solutions in parallel: (1) adding the T5 only when the reaction is already heated to 50°C (using a water bath); (2) using 1/5 of the amount of T5 exo (in both in parallel)
- These are all done in duplicates, resulting in 8 Gibson reactions and subsequent transformations
- Results: (Sunday) There are 2 colonies on one of the plates without either treatment
Sunday (19/08/12)
Ratiometrica-Flu: 3rd attempt at PCR of fragments
- Similar PCR to the one from yesterday, except Velocity mastermix is further changed, and extension time is changed
- Results: failed- no bands, probably Velocity mastermix problem, will revert back to Phusion on next attempt
Week 9
Monday (20/08/12)
Ratiometrica-Flu: 4th attempt at PCR of fragments
- Only vectors this time as we are still not sure if we have got the Velocity mastermix recipe right, as we are waiting for our orders of Phusion to arrive
- Results: no bands for any lanes including positive control
Ratiometrica-Flu: Restreak of potential Ratiometrica colonies
- One of the 2 colonies from the Gibson reaction and subsequent transformation are restreaked on a Amp 100ug/ml plate
- this is incubated 37°C overnight
Wednesday (22/08/12)
Ratiometrica-Lux: PCR of lux-vector
- Subsplit primers for the other half of the fragment that did not come out last week arrived, also new, extended lux FWD primers as we were afraid that the original primers did not anneal properly
- We tried combinations of the new and old primers together with the subsplit and split primers
- Results: only Fragment A2 (Between subsplit FWD and split REV) worked
Ratiometrica-Flu: 5th attempt of PCR of fragments
- Since we got our PCR protocol working, we reattempted the PCR
- Results: the smallest fragments (B0015, K143053) came out, gel extraction is done with the new concentrated protocol
Week 12
Wednesday (12/09/12)
Ratiometrica - lux
- DNA 2.0 construct arrived
- Colonies restreaked and put into 30 °C incubator.
- Only one set of colonies appears to be fluorescing, which may be of concern.
Friday (14/09/12)
PCR of α and β fragments for ratiometrica
- Although ratiometrica appears to have produced successful colonies, we would still like some extra fragments in case we need to repeat our construction.
- Previous successful gel extractions of α and β used as template for PCR.
- α has successfuly been amplified, but &beta produced band of the wrong size.
- α successfully extracted from gel.
Saturday (15/09/12)
Ratiometrica - lux
- Attempted digestion of DNA 2.0 pure plasmid with EcoRI and SpeI, expected fragments 2648bp and 8738bp.
- Parallel digestion of pSB1C3 backbone ready for ligation
- 8738 band extracted, was a little weak
- 8738 band and digested backbone ligated together
- Chemically competent E. coli used for transformation
- Plated on LB Agar with 25μg/ml Chlor
Sunday (16/09/12)
Ratiometrica - lux
- Picked colonies from ligation attempt from yesterday, some red colonies on plates, these were avoided with one exception as a control.
- Picked colonies grown in LB with 25μg/ml Chlor
Week 13=
Monday (17/09/12)
Ratiometrica - Lux
Testing of the 2.0 construct:
Numbers 33,35,36 have correct sequence as of being sent to us. However they were not visibly luminescent. *34 was.
Overnight cultures of each of the 6 culture lines sent were set up. Additionally, inducing plates were streaked, including the 2010 lux biobrick as a luminescence control and pjs130 as an iptg induction control.
Primers ordered to ligate 2.0 construct into pjs130 for insertion into bacillus.
Tuesday (18/09/12)
Ratiometrica - Lux
None of restreaked plates visibly luminescent, aside from *34 (faint), even under photon counting camera. Pjs130 not green, so iptg induction potentially didn't work. LuxBB induced and luminescent.
Cultures induced with 1mM iptg or 3mM arabinose for luxBB, imaged after 3 hrs. Some extremely faint luminescence for 33,34,38 on photon counting camera.
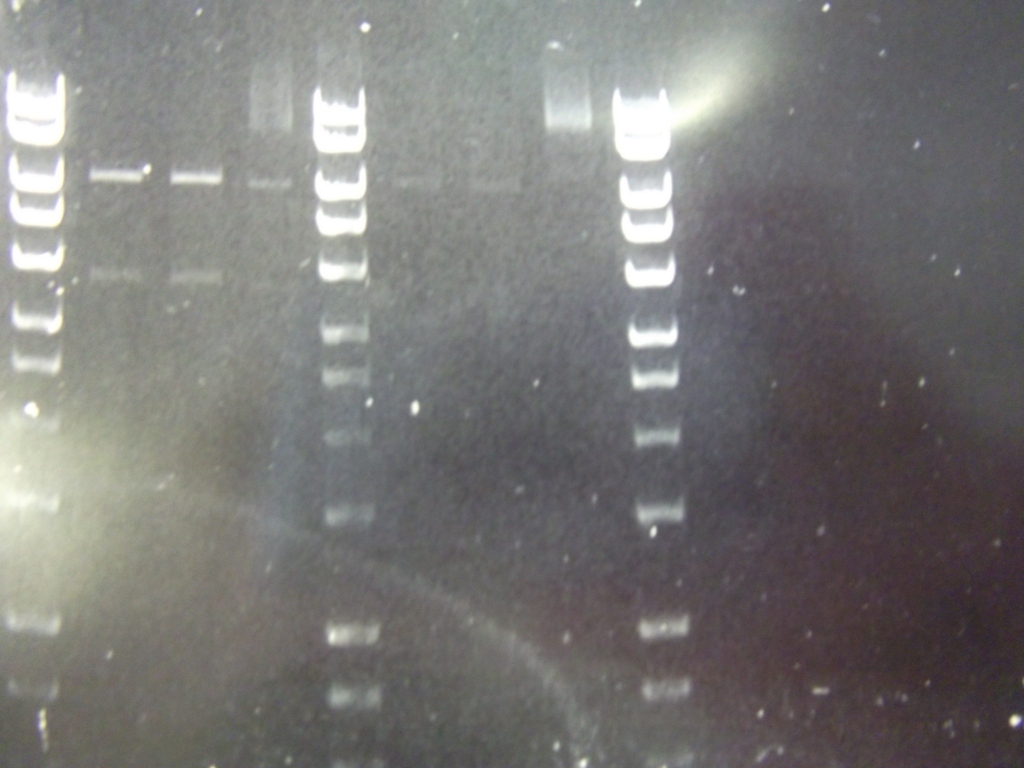
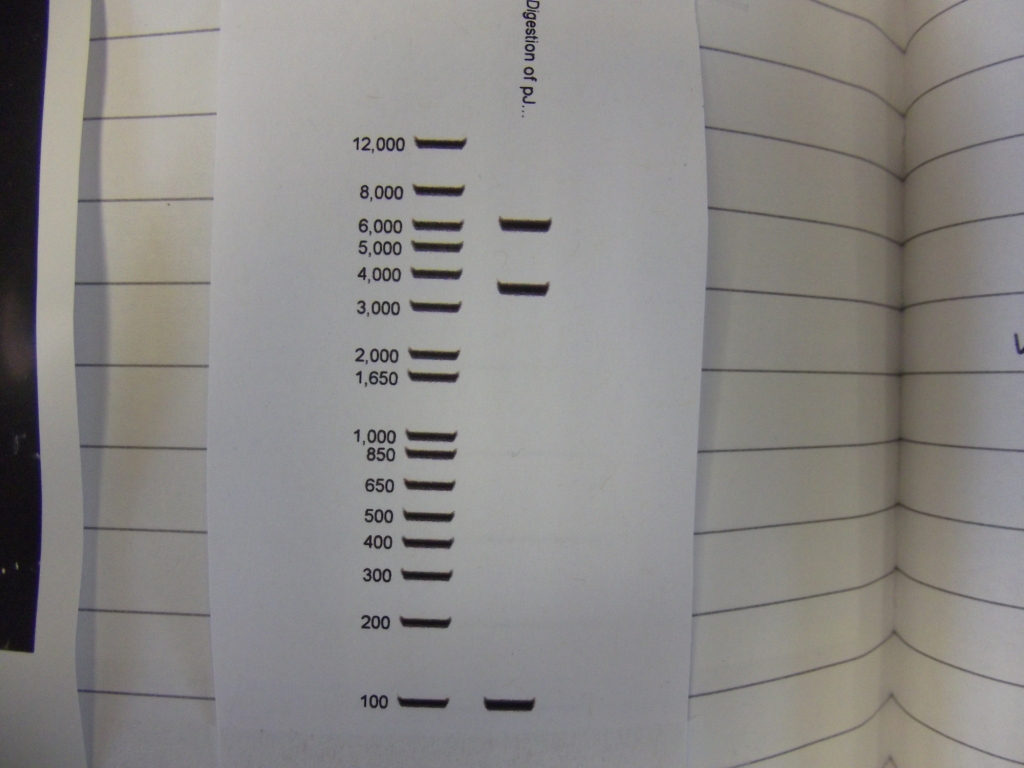
Cultures miniprepped and restriction digests carried out w/spa1. Results are shown. Results indicate problem with the miniprep and/or damage to the construct - expected bands at 6625, 2838, 1876 and 53 bp.
Wednesday (19/09/12)
Ratiometrica - Lux
Digest of 2.0 miniprep with different enzyme, hindIII, carried out. 15ul template DNA used in a 20ul reaction. Again, results were odd. Cut and uncut minipreps ran.Cultures of ratiometrica (fluorescent) were miniprepped to be transformed into bacillus. A restriction digest was performed on these minipreps, and produced bands of the correct size.
Thursday (20/09/12)
Ratiometrica - Lux
Transformation of fluorescent ratiometric construct into bacillus was unsuccessful
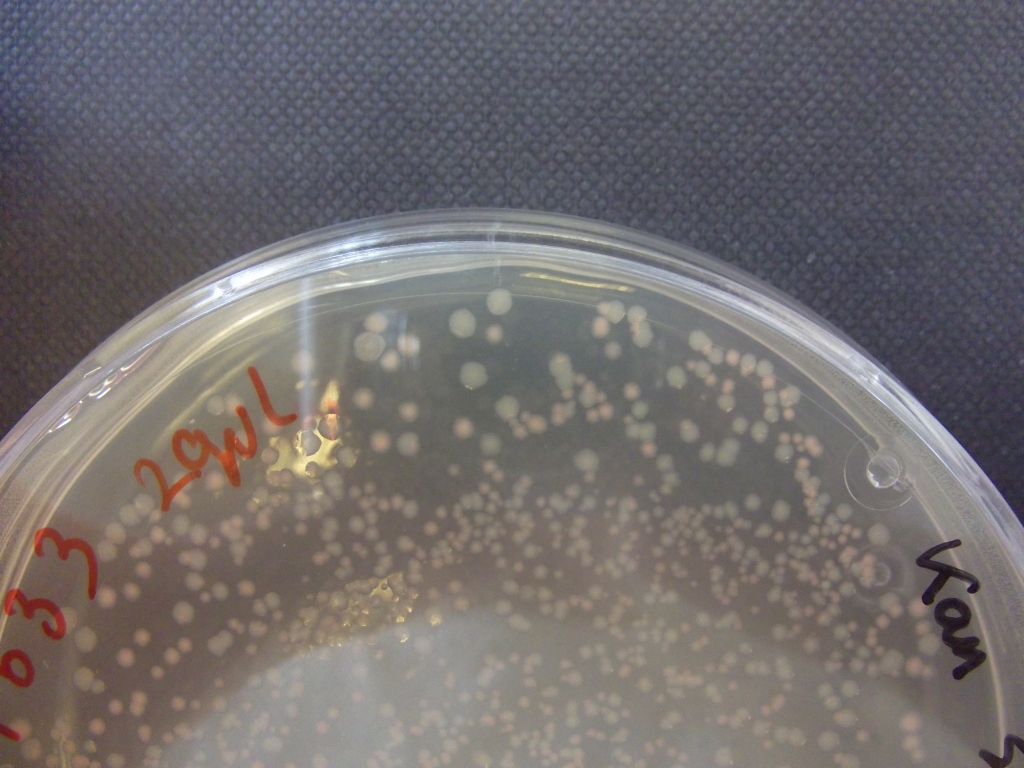
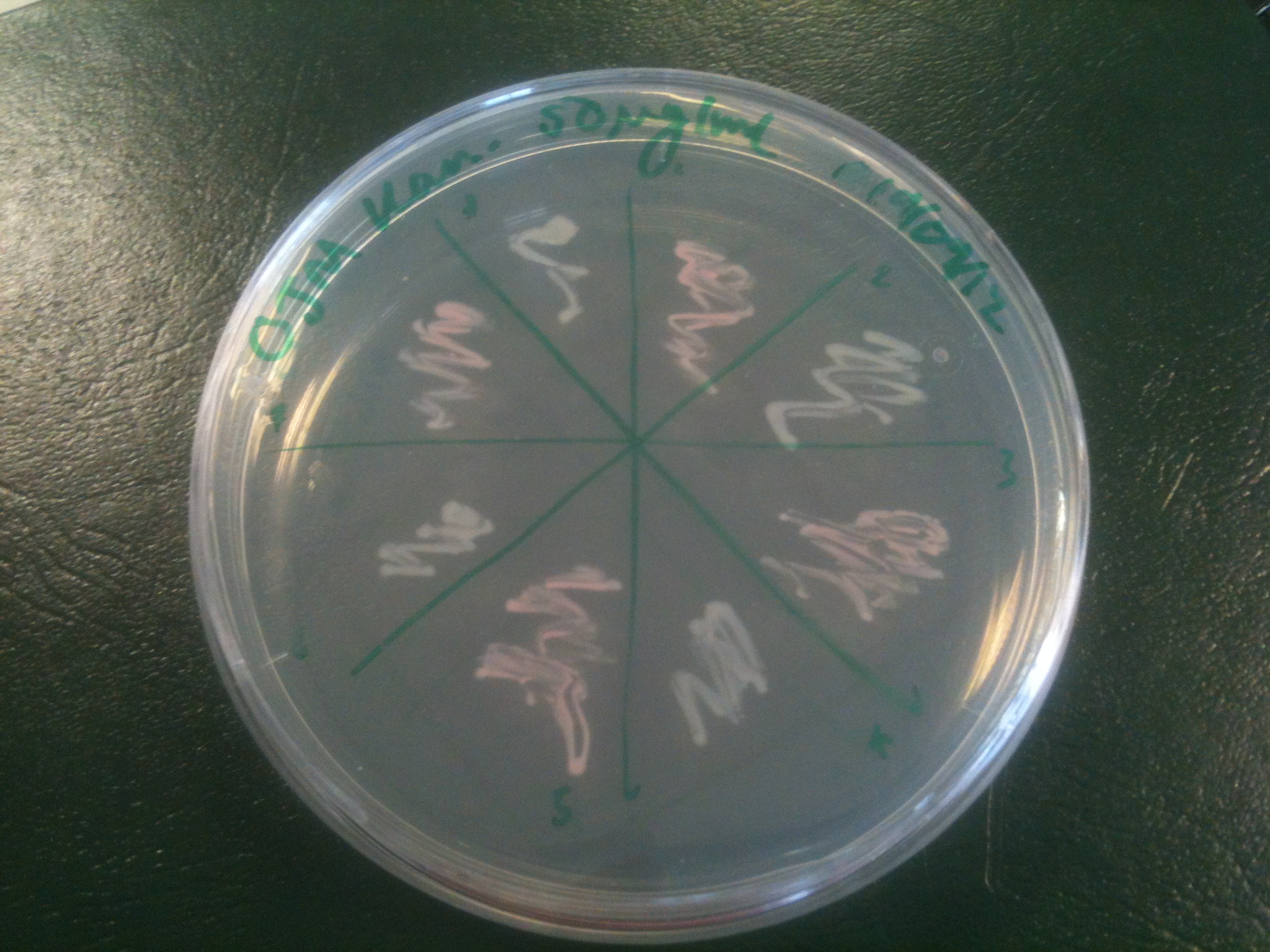
Transformations of XL1-blue cells with resuspended construct plasmid from DNA 2.0 yielded orange colonies, with some white, and some showing sectoring, indicating insert loss.
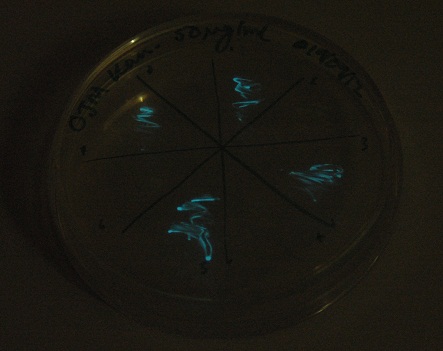
Plates all appear to be luminescent. A random selection of colonies was picked and patched in order to investigate whether luminescence cosegregated with orange colour.
Friday (21/09/12)
Initial pick and patch experiments showed cosegregation, but we did not have immediate access to a photon-counting camera or a DSLR capable of long exposures, so we did not obtain images. Representative colonies were streaked out onto a fresh plate to be imaged later.
Saturday (22/09/12)
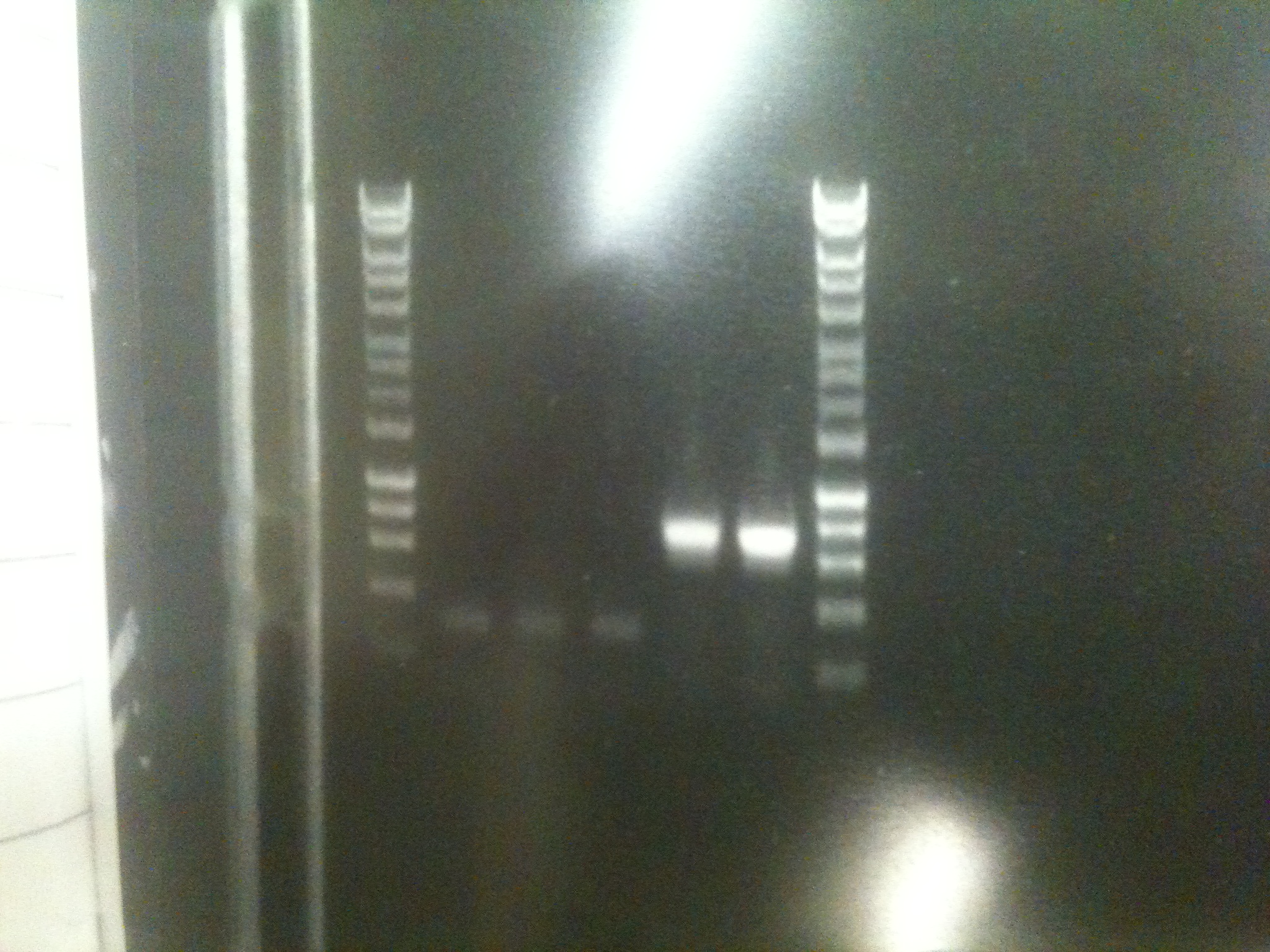
A PCR was run to amplify several fragments, including:
- The ratiometric luciferase construct, to insert into;
- pSB3C5, a low copy number vector
- Oli's Mg riboswitch construct
All fragments were obtained.
Sunday (23/09/12)
Fragments for the fluoride riboswitch and the insertion of the 2.0 construct into pSB3C5 were all digested for two hours in 50 ul reactions, as was the pSB1C3 backbone.
- 5ul NEB2
- 1 ul EcoRI
- 1 ul PstI
- 0.5 ul BSA
- 10 ul DNA
- 32.5 ul HPLC H20
Enzyme was heat-inactivated for 20 minutes at 65 C
for a 6:1 insert:vector ratio (recommended by Knight on openwetware) 2.5 ul of vector digest was ligated to 15 ul insert in all cases, except the luciferase construct. In this case the insert is bigger than the vector, so the ratio was reversed.
- 3ul vector
- 18ul insert
- 2.5ul 10X T4 ligase buffer
- 1.25 ul T4 ligase
Ligation left overnight at 16C, as recommended by Knight.
Week 14
Monday (24/09/12)
Ratiometrica - Lux
Colonies grew. However colony PCR using the sequencing primers revealed them to be transformed with recircularised backbone.The digestions and ligations were reattempted, this time treating the backbone with alkaline phosphatase in order to reduce or eliminate self-ligation.
Tuesday (25/09/12)
Ratiometrica - Lux
Images were taken to show cosegregation of orange colour and luminescence. Additionally, images were taken of the luciferase construct with a gel filter in order to indicate if there was a difference in the emission spectra.
See results page for more details and interpretation.
 "
"