Team:Cambridge/Safety
From 2012.igem.org
Contents |
Judging Form
- Please help the judges by filling out this form. Tell them what medal you think you deserve and why. Tell them which special prizes you should win. Help them find your best parts. Show them how you thought about the safety of your project. Helping the judges will help you too.
- Team: Cambridge
- Region: Europe
- iGEM Year:2012
- Track:Foundational Advance
- Project Name:Parts for a reliable and field ready biosensing platform
- Project Abstract: Implementation of biosensors in real world situations has been made difficult by the unpredictable and non-quantified outputs of existing solutions, as well as a lack of appropriate storage, distribution and utilization systems. This leaves a large gap between a simple, functional sensing mechanism and a fully realised product that can be used in the field.
We aim to bridge this gap at all points by developing a standardised ratiometric luciferase output in a Bacillus chassis. This output can be linked up with prototyped instrumentation and software for obtaining reliable quantified results. Additionally, we have reduced the specialized requirements for the storage and distribution of our bacteria by using Bacillus' sporulation system. To improve the performance of our biosensing platform we have genetically modified Bacillus’ germination speed. Lastly, we demonstrated the robustness of our system by testing it with a new fluoride riboswitch, providing the opportunity to tackle real life problems.
iGEM Medals for non-software teams
- We believe our team deserves the following medal:
- Bronze
- Silver
- √Gold
Because we met the following criteria (check all that apply and provide details where needed)
Requirements for a Bronze Medal
- √Register the team, have a great summer, and plan to have fun at the Regional Jamboree.
- √Successfully complete and submit this iGEM 2012 Judging form.
- √Create and share a Description of the team's project using the iGEM wiki and the team's parts using the Registry of Standard Biological Parts.
- √Plan to present a Poster and Talk at the iGEM Jamboree.
- √Enter information detailing at least one new standard BioBrick Part or Device in the Registry of Standard Biological Parts. Including:
- √Primary nucleaic acid sequence
- √Description of function
- √Authorship
- Safety notes, if relevant.
- √Acknowedgment of sources and references
- √Submit DNA for at least one new BioBrick Part or Device to the Registry.
Additional Requirements for a Silver Medal
- √Demonstrate that at least one new BioBrick Part or Device of your own design and construction works as expected; characterize the operation of your new part/device.
- √Enter this information and other documentation on the part's 'Main Page' section of the Registry
Part Number(s): BBa_K911004
Additional Requirements for a Gold Medal: (one OR more)
- Improve an existing BioBrick Part or Device and enter this information back on the Experience Page of the Registry.
Part Number(s): None - √Help another iGEM team by, for example, characterizing a part, debugging a construct, or modeling or simulating their system.
Link to this information on your wiki. Page name: Team:Cambridge/Outreach/Collaboration - √Outline and detail a new approach to an issue of Human Practice in synthetic biology as it relates to your project, such as safety, security, ethics, or ownership, sharing, and innovation.
Link to this information on your wiki.
Page name: Team:Cambridge/HumanPractices/Overview,Team:Cambridge/HumanPractices/MarketResearch,Team:Cambridge/HumanPractices/FutureDirections
iGEM Prizes
All teams are eligible for special prizes at the Jamborees. more... To help the judges, please indicate if you feel you should be evaluated for any of the following special prizes:
- √Best Human Practice Advance
- √Best Experimental Measurement
- Best Model
Please explain briefly why you should receive any of these special prizes:
Best Human Practice Advance:
We feel that we deserve this prize for three reasons:
- We explored the impacts, *both positive and negative*, of synthetic biology as a solution to real world problems, through interviewing professionals working in a relevant field, namely the impact of arsenic water contamination in Bangladesh.
- We recognized existing problems with the way the current direction of synthetic. On going through the registry we found that most of the characterization data for biosensing parts is often neither comparable nor replicable. We have worked to solve this issue, for example with our ratiometric dual channel output.
- *Our project doesn’t stop here*, in Chanel number 6 (Team:Cambridge/HumanPractices/FutureDirections) we considered the future implications and technological applications of our project, as well as the means by which it could be improved by subsequent users. We feel that the end to an iGEM project should not be the conclusion of an idea, but the start of it.
Best BioBrick Measurement Approach:
It is absolutely vital that a quantitative, numerical, robust, and flexible measurement approach exists to relay information to a user that is an accurate representation of the input processed by a biological device. Working from these principles, the following was done:
- We designed and built Biologger, a *cheap, arduino-based, fully functional automatic rotary device* that has an incorporated ratiolumnometer
- Our project is entirely open-sourced and open-platform. We have published source code for the two applications which serve to operate the device, one for PCs and the other for Android devices, as well as the open source circuit design that provides this ratiometric reading. Furthermore, the Android app is able to receive its data wirelessly, which we feel is a great advance in BioBrick measurement.
- Our dual-channel luciferase reporter was successfully tested with a dilution series of E.coli transformed with the Lux Operon (under pBAD) biobrick (Part BBa_K325909) of the Cambridge iGEM 2010 team. It can detect, with good accuracy, both different light intensities, as well as the percentages of blue or orange frequencies in a sample.
- Our device was successfully tested using artificial light to detect different frequencies (colours) as well.
Having done all the above, we believe that this fully open-sourced instrumentation kit (mechanical) chassis, electronics, software code), estimated at *$35.00* (or $85.00 if a Bluetooth modem is required), is a complete BioBrick measurement solution for any and all BioBricks with a light output.
Team_Parts
To help the judges evaluate your parts, please identify 3 of your parts that you feel are best documented and are of the highest quality.
- Best new BioBrick part (natural)
- BBa_K911003
- Best new BioBrick part (engineered)
- BBa_K911004
- Best improved part(s): None
List any other parts you would like the judges to examine:BBa_K911001, BBa_K911009, BBa_K911008
Please explain briefly why the judges should examine these other parts:
- Magnesium Sensitive Riboswitch BBa_K911001
As a riboswitch sensing construct, this part is an entirely new type of biosensor (along with the fluoride construct) that could potentially change the way we think about designing input genetic circuits. Unlike the fluoride riboswitch, it is a derepression system and therefore serves to demonstrate the principle that riboswitches can be used regardless of whether they turn on or off their reporter. - Fluorescent ratiometric construct for standardizing promoter output BBa_K911009
Fluorescence is a major cornerstone for biosensors in the registry, however, most parts do not involve the use of a ratiometric output, which has been shown in the literature to provide much more reliable and meaningful data. This part not only furthers the development of ratiometric measurements in molecular biology but due to the choice of promoters and terminators it can be used to characterize the difference in activity between E. coli and B. Subtilis - Fast Germination (B. subtilis) BBa_K911008
This part is entirely novel for the registry and fully utilizes the recombination machinery inherent in the Bacillus chassis. Have spores that can germinate at a faster rate is certainly a worthy achievement and could help with experiments with B. Subtilis that any future iGEM teams may wish to perform.
iGEM Safety
For iGEM 2012 teams are asked to detail how they approached any issues of biological safety associated with their projects.
The iGEM judges expect that you have answered the four safety questions Safety page on your iGEM 2012 wiki.
Please provide the link to that page: Page name: Team:Cambridge/Safety
Attribution and Contributions
For iGEM 2012 the description of each project must clearly attribute work done by the team and distinguish it from work done by others, including the host labs, advisors, and instructors.
Please provide the link to that page, or comments in the box below: Page name: Team:Cambridge/Attributions
Comments
If there is any other information about your project you would like to highlight for the judges, please provide a link to your wiki page here: Team:Cambridge/Overview/DesignProcess
Safety
N.B. This page is a work in process and will be added to and improved over the course of the project.
iGEM safety questions
1. Would any of your project ideas raise safety issues in terms of:
- researcher safety,
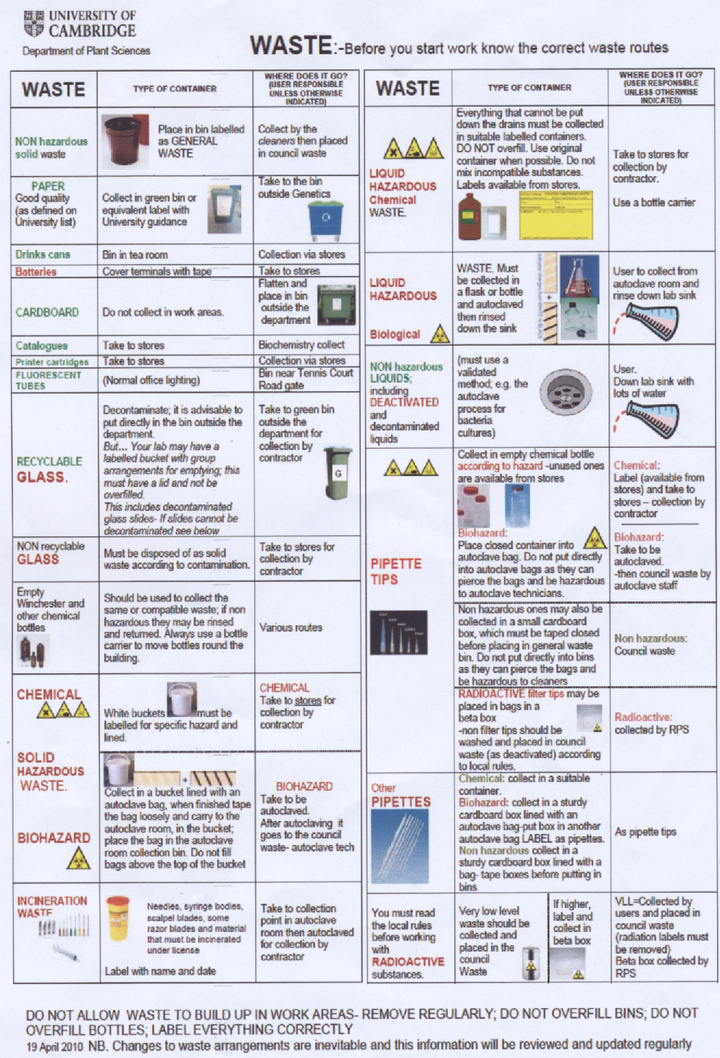
Our laboratory is only authorised to use Biosafety level 1 (non-pathogenic) bacteria and we therefore consider our project to be harmless to researchers as none of our experiments have any known action in increasing the pathogenicity of the bacteria we use or the host range. It is departmental policy however to treat all bacteria as potential pathogens and to take suitable safety precautions. We understand that it is essential to wear suitable PPE (personal protective equipment) at all times, in our experiments this usually means labcoats and gloves. All laboratory equipment and bacterial cultures are to be decontaminated by autoclaving. A copy of the departmental regulations for waste disposal is available at the bottom of this page.
Several techniques used in the course of this project call for the use of potentially hazardous substances which range from causing minor skin irritations to being toxic if ingested. For this reason we have created risk assessment pages for each of the protocols used in the course of our research. These are not intended to be used instead of departmental safety procedures, but outline the major concerns for each protocol for reference by future teams. MSDS sheets for reagents used in the course of our project are also available on this wiki.
Sodium Fluoride is hazardous and was necessary for testing our fluoride riboswitch. Full departmental risk assessments were completed before working with this substance and a brief risk assessment outlining the main dangers can be found on this wiki (here) as well as a MSDS.
We anticipate that elements of our project could be used for a wide variety of purposes and these may include the use of potentially harmful substances. Any future teams would need to consider the safety implications of their project on a case by case basis, though our project should not raise any safety concerns.
- public safety,
We understand public apprehension surrounding the use of genetically modified bacteria, in the unlikely event that members of the public came into contact with the bacteria used in our project we anticipate that there would be no threat to public safety as we are using non-pathogenic strains.
- environmental safety?
While we appreciate that pathogenic strains of the species we are using exist, we are using disabled, non-pathogenic strains that should otherwise interact in an identical manner with the environment. We therefore anticipate that any released bacteria would be disadvantaged and are not expected to survive outside of the favorable conditions engineered in the lab.
Should the goals of our project come to fruition, there are additional safety factors that must be considered upon the implementation of our kit in the field. Our project must be able to remain within the bounds of current laws to be considered a success. Biocontainment of the bacteria used as the biosensors is therefore a high priority. Current systems are typically somewhat cumbersome and unreliable in their implementation - for example, sample tubes may have to be collected and then sterilized with boiling water. In order to ensure that no GMOs are released into the natural environment upon use of our system, we have devised several strategies that could be used to prevent release of viable bacteria from our kit:
- The storage cuvettes could contain a small container of bleach. Breaking a seal when the cuvette is finished with results in its release and causes destruction of the active bacteria. The cuvette can then be disposed of in general waste, with all GMOs inside destroyed.
- During sporulation, a unique sigma factor is activated. This acts upon promotors that bring about the sporulation response. Our cells could therefore be engineered to activate an internal clock upon sporulation that would remain activated after germination and cause the destruction of the cells after a certain number of generations.
- The cells could be engineered to require a complex substance that is only practically available in their medium. Should the cells stray from their medium, an autolysis gene would be activated and the cells would self destruct. This is analogous to the survival factors required by mammalian cells to prevent apoptosis.
Use of several (or, for preference, all) of these techniques should prevent unintentional spread of our GMOs. Taking a lesson from the development of antibiotic resistance in bacterial strains, we hope that with several failsafes such as the ones that we are proposing in place, the likelihood of any single cell surviving the entire series of biocontainment measures will fall to essentially zero.
Please note that we are not currently implementing these strategies. While it is important to consider how we would prevent the spread of GMOs if we were to use them in the field, the measures discussed above will prevent their spread from the research environment.
2. Do any of the new BioBrick parts (or devices) that you made this year raise any safety issues? If yes,
- did you document these issues in the Registery?
- how did you manage to handle the safety issue?
- how could other teams learn from your experience
Our biobricks are designed to create a biological system whereby a bacterial cell takes in some substance to be measured and this induces a light output. None of our biobricks increase either the pathogenicity of the bacteria used or the range of usable hosts.
Our biobricks do not raise any safety issues.
Nevertheless, we are remaining vigilant in ensuring that no GMOs come into contact with either researchers or the wider world.
3. Is there a local biosafety group, committee or review board at your institution?
- If yes, what does your local biosafety group think about your project?
Departmental Codes of Practice for GM organisms were also consulted before we began our project. We have thus been cleared to work with bacteria that have been classified as being "unlikely to cause human disease".
Our project and protocols have been reviewed and accepted by our advisers and the departmental safety officer as suitable.
As we are a UK team there are also national biosafety regulations (here) that we were made aware of before beginning our project. We have complied with all of these guidelines, and hence have worked well within the law.
- If no, which specific biosafety rules or guidelines do you have to consider in your country
N/A
4. Do you have any other ideas how to deal with safety issues that could be useful for future iGEM competitions? How could parts, devices and systems be made even safer through biosafety engineering?
Biosafety was a major consideration when planning our project and many (we feel) quite brilliant project ideas were discarded because they posed significant safety risks or the procedures it would be necessary to implement to work safely were unfeasible within the time scale of the project. Project ideas that would have required the widespread or uncontrolled release of our bacteria into the environment were also discarded.
We recommend that future teams consider following this planning procedure, as the real world applications proposed by many past teams appear to fail to take into consideration the risks posed by the uncontrolled release of their GMOs into the wider environment.
We also recommend that future teams try to consider failsafes in the biocontainment aspects of their projects to reduce the risks of GMOs evolving resistance to biocontainment efforts.
Additional safety information
- Risk Assessments Risk assessments for any experiments we do and safety information for carrying out any of the protocols listed on this wiki.
- MSDS Sheets Materials Safety Data Sheets for the reagents used in the course of our project.
- Protocols Protocols used in the course of our project.
- Waste procedure summary A copy of several waste safety posters around the department:
 "
"