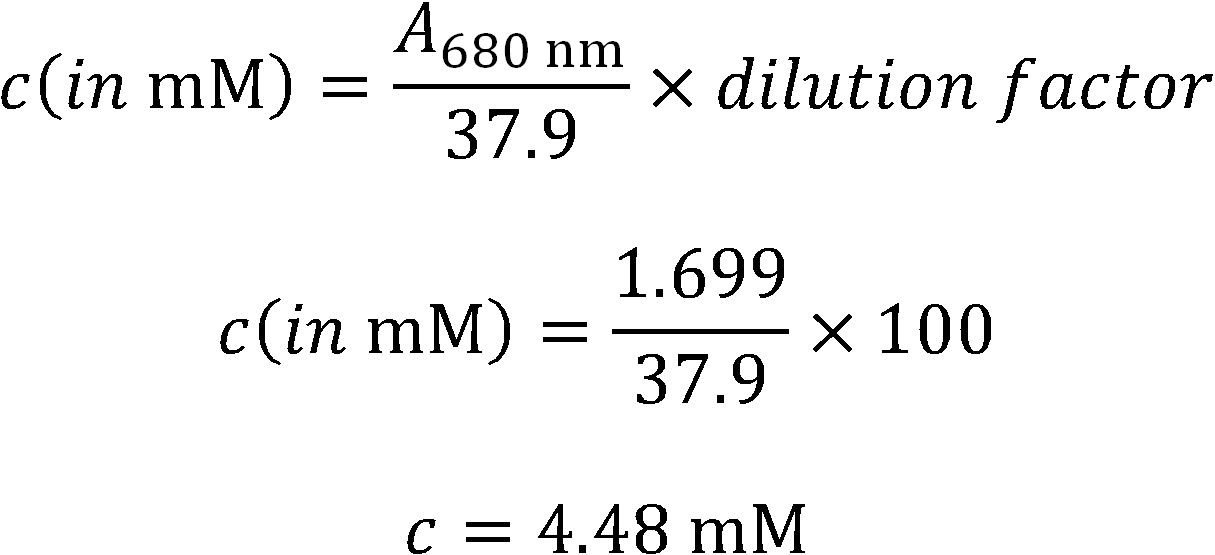Team:TU Munich/Project/Light Switchable Promoter
From 2012.igem.org



Contents |
Light-switchable Promoter
The so-called "Reinheitsgebot" or "Bavarian Beer Purity Law" forbids the use of any ingredients other than water, barley and hops. Hence, to be able to control the expression of our pathways in yeast, a promoter which does not rely on any chemical additive.
The light switchable promoter, does not only comply with these needs, it is also easy, cheap and very precisely applicable. Furthermore, as the expression of the downstream gene can be upregulated as well as downregulated by variation of red light and far red light ratio respectively.
Therefore it allows high spatio-temporal control over the genes downstream of the promoter.
Background and principles
This system bases on the yeast two-hybrid system which was originally created for exploring protein-protein interactions. One candidate of a potential protein-interaction pair is fused to the DNA-binding domain of a transcription factor and the other candidate to the activation domain of a transcription factor. If the proteins candidates are really physically interacting with each other, this event will starts the transcription of downstream reporter genes, e. g. LacZ or an auxotrophic marker.
Reverse yeast-two hybrid
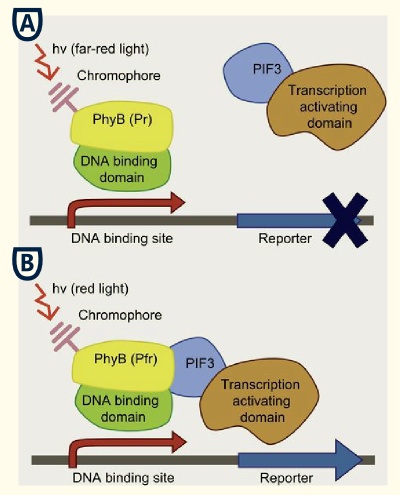
This basic principle is utilized in the yeast light-switchable promoter system. But in contrast to yeast-two hybrid, we already know the interaction partners (PhyB and PIF3). The photoconvertible binding of PhyB to PIF3 is used, to recover the physical contiguity of the DNA binding domain and the transcriptional activation domain under defined conditions (red light).
This light-inducible system contains two proteins, phytochrome B (PhyB) and phytochrome interacting factor 3 (PIF3). PhyB and PIF3 will just form a heterodimer, if PhyB is exposed to red light. Exposition under red light leads to a conformation change of PhyB to its active form (Pfr-form); the Pfr form of PhyB now can bind PIF3. PhyB comprises a light-absorbing chromophore phycocyanobilin, which gives PhyB the ability to undergo a photoconversion to the active Pfr form (red light exposition) or back to its ground-state Pr (far-red light exposition or darkness).
GAL4 based light-switchable promoter system
In our first case we create two constitutively expressed fusion proteins, the first one is PhyB fused to GAL4DBD for the DNA binding part (BBa_K801040 and the second one is PIF3 fused to GAL4AD for the transcriptional activating part (BBa_K801039). This system allows us to control spatio-temporally the expression of our genes coded on pTUM104 and driven by the GAL1 promoter (The TATA-box of pGAL1 is preceded by binding elements for GAL4). To prevent interference with the endogenous GAL4 system of yeast, we are using the Y190 S. cerevisiae strain, which has an GAL4/GAL80 deletion.
LexA based light-switchable-promoter system
In contrast to the GAL4 based light-switchable promoter system there is no need for KO of GAL4/GAL80 genes in yeast with a LexA based light-switchable promoter system. The difference is that we use LexA, a prokaryotic DNA binding protein, for the DNA binding part of our light-switchable promoter system, instead of GAL4DBD. LexA does not interfere with the endogenous yeast metabolism and signalling system because it only recognizes a special prokaryotic DNA sequence, the so-called LexA operator (=LexA binding site). LexA binding sites can be used upstream of a minimal promoter (=TATA box) to be utilized as a cis-acting regulatory element.
In this case the genes, which we want to control by light, have to be cloned downstream of a synthetic promoter containing a minimal promoter, preceded by multiple LexA binding sites, e. g. BBa_K165031.
Biosynthesis of phycocyanobilin
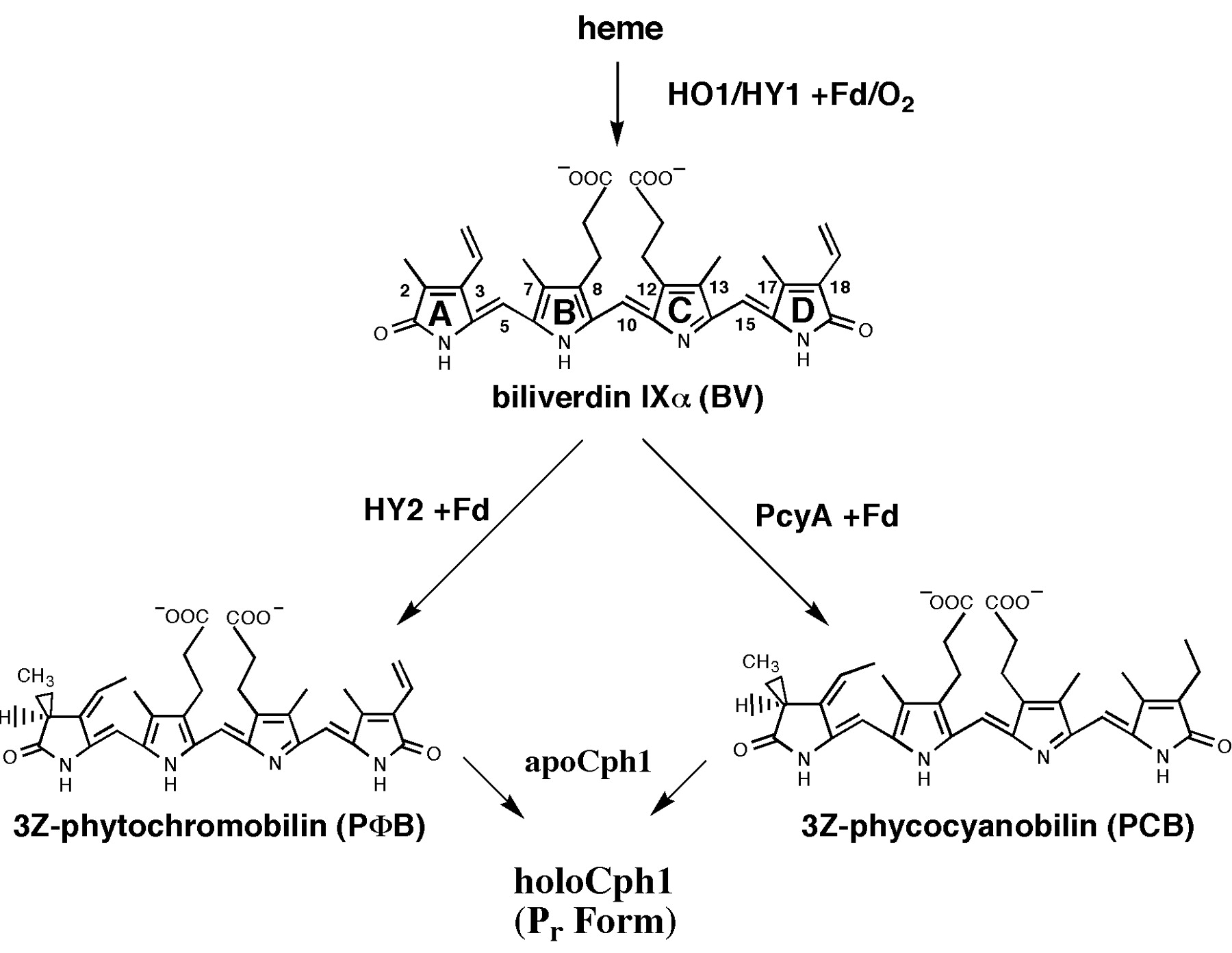
Phycocyanobilin undergoes a Z-E isomerization to its active form in case of red light and an E-Z isomerization to its inactive form in case of far-red light. The half-life of its active form Pfr is ~30 min, so continuous red light exposition is not nescessary. A great adventage is that light-sensitive odorant and flavorings will not be destroyed. Once phycocyanobilin is not naturally avaible in yeast one have to add the tetrapyrrole light-absorbing chromophore phycocyanobilin to the medium to get a functional light-switchable promoter system. But it also possible to bring the capability of phycocyanobilin synthesis in yeast by metabolic engineering. From heme, which is endogenous in yeast, there are only two steps of biosynthesis away from phycocyanobilin. The first step of phycocyanoblin is catalized by a heme oxygenase and the second step by a phycocyanobilin:ferredoxin oxidoreductase.
Results
Components of the light-switchable-promoter systems
There a two fusion proteins nescessary for a light-switchable promoter system. The first one is PIF3 fused to GAL4AD and the second one is GALDBD (GAL4 based) or LexA (LexA based) fused to PhyB.
For PhyB and PIF3 we didn't use the whole protein coding sequence for our fusions. For PhyB we used the first 908 N-terminal amino acids which has been mapped to be sufficient for reversible photoconversion. Also for PIF3 only the first 100 N-terminal aminoa acids has been taken for our fusions because they has been mapped to be only nescessary for light-switchable binding to PhyB.
We successfully created all fusion proteins for a light-switchable promoter system based on GAL4 and LexA and even creates a TEF1 promoter driven expression battery for all our components for each type of the system (GAL4 and LexA based).
- Fusion protein for the first component (GAL4/LexA based):
BBa_K801039: SV40NLS-GAL4AD-Linker-PIF3
- Fusion protein for the second component (GAL4 based):
BBa_K801040: SV40NLS-PhyB-Linker-GAL4DBD
- Fusion protein for the second component (LexA based):
BBa_K801041: SV40NLS-PhyB-Linker-LexA
- TEF1 promoter driven gene expression battery for all parts of the GAL4 based light-switchable-promoter system:
BBa_K801042: pTEF1_SV40NLS-GAL4AD-Linker-PIF3_tTEF1_pTEF1_SV40NLS-PhyB-Linker-GAL4DBD_tTEF1
- TEF1 promoter driven gene expression battery for all parts of the LexA based light-switchable-promoter system:
BBa_K801043: pTEF1_SV40NLS-GAL4AD-Linker-PIF3_tTEF1_pTEF1_SV40NLS-PhyB-Linker-GAL4LexA_tTEF1
Components for reporters
GAL4 based reporter systems
For the GAL4 based light-switchable promoter system we have endogenous reporters in the Y190 S. cerevisiae strain.
The first one is an auxotrophic reporter for HIS3, an imidazoleglycerol-phosphate dehydratase, which catalyses the sixth step in histidine biosynthesis. HIS3 is driven by a synthetic promoter with upstream GAL4 responsive elements. If plated or inoculated on/in histidine deficient medium, there should be no growth of yeast if they incubated in darkness or far-red light conditions. But under red light conditions the auxotrophy is reverted by expression of HIS3 due to the recruitment of GAL4AD through PhyB-PIF3 interaction.
The second reporter is LacZ, a beta-galactosidase, which is under the control of pGAL1. Beta-galactosidase is only expressed, when the light-switchable promoter system is switched on by red light.
LexA based reporter systems
For the LexA based light-switchable promoter system we have to transfect yeast with a second plasmid coding for the reporter construct because there is no endogenous reporter system like for the GAL4 based system. Also we didn't use the GAL4/GAL80 deletion strain Y190 in contrast to the GAL4 based system since there is no need for that because there is no interference by the prokaryotic LexA system with the endogenous yeast signalling and metabolism pathways.
We've successfully cloned a luciferase from Renilla reniformis (BBa_J52008) downstream of a minimal CYC1 promoter preceded by LexA binding sites (BBa_K165031).
Extraction of PCB
Since there is no endogenous phycocyanobilin (PCB) in yeast, we have to add it to the medium first for our first proove-of-concept experiments. Later, we can implement the enzymes for the biosynthesis of phycocyanobilin (BBa_I15008 and BBa_K181000) also in the finished gene expression batteries for our light-switchable promoter systems(BBa_K801042 and BBa_K801043).
- Phycocyanobilin is extracted by methanolysis of dried Spirulina platensis. For detailed information please see our methods section
- The extracted phycocyanobilin is resuspended in DMSO and is kept at -20 °C until use.
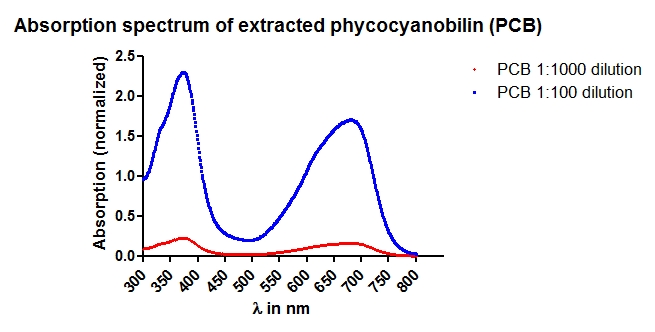
- Absorption Spectrum for concentration determination.
 "
"