Team:TU Munich/Project/Light Switchable Promoter
From 2012.igem.org



Contents |
Light-Switchable Promoter
The so-called "Reinheitsgebot" or "Bavarian Beer Purity Law" forbids the use of any ingredients other than water, barley and hops. Hence, to be able to control the expression of our pathways in yeast, a promoter which does not rely on any chemical additive is needed.
The light switchable promoter, does not only comply with these needs, it also is easy, cheap and very precisely applicable. Furthermore, as the expression of the downstream gene can be upregulated as well as downregulated by variation of red light and far red light ratio respectively.
Hence it allows high spatio-temporal control over the genes downstream of the promoter.
Background and principles
This system bases on the yeast two-hybrid system which was originally created for exploring protein-protein interactions. One candidate of a potential protein-interaction pair is fused to the DNA-binding domain of a transcription factor and the other candidate to the activation domain of a transcription factor. If the proteins candidates are really physically interacting with each other, this event will starts the transcription of downstream reporter genes, e. g. LacZ or an auxotrophic marker.
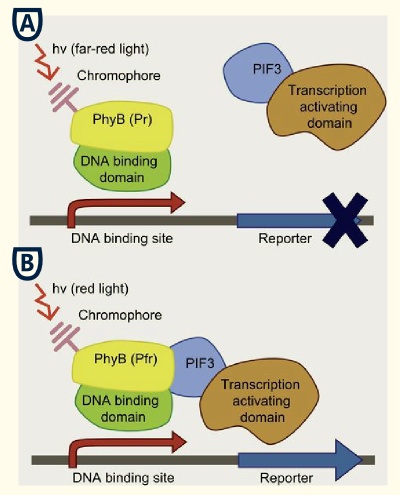
This basic principle is utilized in the yeast light-switchable promoter system. But in contrast to yeast-two hybrid, we already know the interaction partners (PhyB and PIF3). The photoconvertible binding of PhyB to PIF3 is used, to recover the physical contiguity of the DNA binding domain and the transcriptional activation domain under defined conditions (red light).
This light-inducible system contains two proteins, phytochrome B (PhyB) and phytochrome interacting factor 3 (PIF3). PhyB and PIF3 only form a heterodimer together when PhyB is exposed to red light. Exposition under red light leads to a conformation change of PhyB to its active form (Pfr-form); the Pfr form of PhyB now can bind PIF3. PhyB consists a light-absorbing chromophore phycocyanobilin, which gives PhyB the ability to undergo a photoconversion to the active Pfr form (red light exposition) or back to its ground-state Pr (far-red light exposition or darknes).
Phycocyanobilin undergoes a Z-E isomerization to its active form when exposed to red light and an E-Z isomerization to its inactive form when exposed to far-red light. The half-life of its active form Pfr is ~30 min, so no continuous red light exposition is nescessary. Once phycocyanobilin is not naturally avaible in yeast one have to add the tetrapyrrole light-sbsorbing chromophore phycocyanobilin to the medium to get a functional light-switchable promoter system. But it also possible to bring the capability of phycocyanobilin synthesis in yeast by metabolic engineering. From heme, which is endogenous in yeast, there are only two steps of biosynthesis away from phycocyanobilin. The first step of phycocyanoblin is catalized by a heme oxygenase and the second step by a phycocyanobilin:ferredoxin oxidoreductase.
Idea
General remarks and issues
- A yeast strain with GAL4/GAL80 deletion is required to avoid interference by endogenous GAL4 and GAL80 proteins. Disadvantage of GAL4/GAL80 deletion is that the cells are growing more slowly compared to strains with the wildtype alleles of these genes. Use of prokaryotic LexA instead of the DNA-binding domain of GAL4 avoids that. In this case the UAS (upstream activation sequence) of GAL1 has to be replaced by a prokaryotic upstream LexA operator.
- For a perfect light-switchable system I need an idea for inactivating/degrading LacI fastly because the half-life of LacI is about 10 min. Is it fast enough?
- Check for avaible Nuclear Localization Signals (NLS) for nuclear localized proteins!
- Better we synthesize the DNA of some promoters including UAS and operators.
- The first ~650 N-terminal amino acids are only needed for a functional PhyB.
- The first ~100 N-terminal amino acids are only needed for a fucntional Pif3.
- I asked Roman, if he can get all the elements for Yeast-two-hybrid-system including the GAL4/GAL80 deletion strain from Professor Schwab. May be he also has the modifed Yeast-two-hybrid-sytem with LexA.
Results
Extraction of PCB
- Short Description of the procedure
- Picutere of the finished extract
- Absorption Spectrum for concentration
 "
"