Team:TU Munich/Project/Ethanol Inducible Promoter
From 2012.igem.org
Simon Heinze (Talk | contribs) (→Idea) |
Simon Heinze (Talk | contribs) (→Second experiment: Characterization of the KlADH4-promoter without over-night pre-cultures) |
||
| (36 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
[[File:Simon_einzel_TUM12.jpg|200px|thumb||Responsible: Simon Heinze]] | [[File:Simon_einzel_TUM12.jpg|200px|thumb||Responsible: Simon Heinze]] | ||
| - | An ethanol inducible promoter - that would really be handy in brewing a beer with transgenic yeast! According to the German Purity Law, no ingredients other than water, hops and barley may be used for brewing - which is why we can't simply add a chemical to induce gene expression at the desired point of time. But constitutive promoters are not always the best solution, so we thought about substances that "appear" during brewing. And of course we came up with - Alcohol! Alcohol would be the ideal inducing agent because the yeast cells could take care of the main fermentation first, without any heterologous pathways hindering them. Then, after the main fermentation is completed | + | An ethanol inducible promoter - that would really be handy in brewing a beer with transgenic yeast! According to the German Purity Law, no ingredients other than water, hops and barley may be used for brewing - which is why we can't simply add a chemical to induce gene expression at the desired point of time. But constitutive promoters are not always the best solution, so we thought about substances that "appear" during brewing. And of course we came up with - Alcohol! Alcohol would be the ideal inducing agent because the yeast cells could take care of the main fermentation first, without any heterologous pathways hindering them. Then, after the main fermentation is completed, ethanol is present and induce the heterologous genes - the yeast produces our desired substances. |
| - | Another application for an alcohol inducible promoter is that of a classic biosensor. If cloned in front of a | + | Also beyond the scope of brewing an ethanol sensitive promoter would have a broad spectrum of application. If the induction of heterologous gene expression occurs at sufficiently high ethanol concentrations, a system similar to the T7-promoter based auto-induction system in ''E. coli'' (Studier, 2005) could be established. |
| + | |||
| + | Another application for an alcohol inducible promoter is that of a classic biosensor. If cloned in front of a reporter gene such as GFP, the fluorescence could be used to deduce the current ethanol content, thus abolishing the need for complicated enzyme assays or distillation experiments, which are currently among the most applied tools for measuring the alcohol content in beverages. | ||
We chose the KlADH4-promoter because it originates from a close relative of ''S. cerevisiae'' - ''Kluyveromyces lactis''. It has been reported that promoters can be interchanged between those two species and in addition to that, all transcription factors and cis-elemtents known to be involved in the alcohol response of the KlADH4 promoter in ''K. lactis'' also occur in ''S. cerevisiae'' (Mazzoni et al., 2000). Because of that, we do not need to express genes for any additional transcription factors. | We chose the KlADH4-promoter because it originates from a close relative of ''S. cerevisiae'' - ''Kluyveromyces lactis''. It has been reported that promoters can be interchanged between those two species and in addition to that, all transcription factors and cis-elemtents known to be involved in the alcohol response of the KlADH4 promoter in ''K. lactis'' also occur in ''S. cerevisiae'' (Mazzoni et al., 2000). Because of that, we do not need to express genes for any additional transcription factors. | ||
| Line 23: | Line 25: | ||
'''KlADH4''' | '''KlADH4''' | ||
| - | [[File: | + | [[File:TUM12_Sequence_KlADH4_prom.png|thumb|lright|280px|Nucleotide sequence of the KlADH4 promoter region responsive to ethanol induction (UAS<sub>E</sub>). The five STREs (shadowed boxes), the putative HSEs, the RapI and the YapI binding sites are also indicated. Nucleotides matching with the HSE consensus sequence are underlined. Picture taken from: Mazzoni et al., 2000]] |
| - | The ''K. lactis'' gene KlADH4 encodes a mitochondrial | + | The ''K. lactis'' gene KlADH4 encodes a mitochondrial alcohol dehydrogenase which can be specifically induced by ethanol and is insensitive to glucose repression. A small region in the KlADH4 promoter, called UAS<sub>E</sub>, renders the expression of a gene ethanol-dependent (Mazzoni et al., 2000 [http://www.ncbi.nlm.nih.gov/pubmed?term=10724480]). UAS<sub>E</sub> is a cis-element of the KlADH4 promoter spanning from -953 to -741. When inserted into the promoters of heterologous genes, these genes become ethanol-inducible in ''K lactis''. |
The sequence contains (Mazzoni et al. 2000): | The sequence contains (Mazzoni et al. 2000): | ||
| Line 32: | Line 34: | ||
*consensus sequence of the binding site of Rap1 protein. RAP1 (repressor activator protein 1) is an essential gene present in both ''S. cerevisiae'' and ''K. lactis'' and acts positively or negatively on the expression of many genes. Rap1 binding sites have been found in promoters of glycolytic genes where they play a positive role in transcription. | *consensus sequence of the binding site of Rap1 protein. RAP1 (repressor activator protein 1) is an essential gene present in both ''S. cerevisiae'' and ''K. lactis'' and acts positively or negatively on the expression of many genes. Rap1 binding sites have been found in promoters of glycolytic genes where they play a positive role in transcription. | ||
*Five STREs (stress response elements). STREs have been found in the promoters of many ''S. cerevisiae'' genes, where they are present in two or more copies in both orientations | *Five STREs (stress response elements). STREs have been found in the promoters of many ''S. cerevisiae'' genes, where they are present in two or more copies in both orientations | ||
| - | *Two HSEs (heat shock elements). HSEs have been found in combination with STREs in many stress-responsive genes of S. cerevisiae. | + | *Two HSEs (heat shock elements). HSEs have been found in combination with STREs in many stress-responsive genes of ''S. cerevisiae''. |
| - | Despite the presence of the STREs, KlADH4 is '''not''' a stress responsive gene but specifically induced by | + | Despite the presence of the STREs, KlADH4 is '''not''' a stress responsive gene but specifically induced by alcohol (Mazzoni et al., 2000). |
</div> | </div> | ||
| Line 40: | Line 42: | ||
<hr/> | <hr/> | ||
<div> | <div> | ||
| - | Although there is evidence that the KlADH4 promoter can be used for ethanol dependent production of recombinant proteins in ''K. lactis'' ([http://www.ncbi.nlm.nih.gov/pubmed/9872759 Saliola et al. 1999]), we did not find any literature which indicated that this alcohol- | + | Although there is evidence that the KlADH4 promoter can be used for ethanol dependent production of recombinant proteins in ''K. lactis'' ([http://www.ncbi.nlm.nih.gov/pubmed/9872759 Saliola et al. 1999]), we did not find any literature which indicated that this alcohol-inducible promoter has been used in other organisms such as ''S. cerevisiae''. |
Mazzoni et. al (2000) annotated binding sites for two transcription factors (Rap1 and Yap1) as well as some small cis-elements (HSEs and STREs) in the UAS<sub>E</sub>-region of the KlADH4-promoter. All of these factors are not only present in ''K. lactis'' but also in ''S. cerevisiae'' (Kuge & Jones, 1994 [http://www.ncbi.nlm.nih.gov/pubmed?term=8313910], Shore & Nasmyth, 1987 [http://www.ncbi.nlm.nih.gov/pubmed?term=3315231], Schueller et al., 1994 [http://www.ncbi.nlm.nih.gov/pubmed?term=7523111], Ruis & Schueller, 1995 [http://www.ncbi.nlm.nih.gov/pubmed?term=8526890]). | Mazzoni et. al (2000) annotated binding sites for two transcription factors (Rap1 and Yap1) as well as some small cis-elements (HSEs and STREs) in the UAS<sub>E</sub>-region of the KlADH4-promoter. All of these factors are not only present in ''K. lactis'' but also in ''S. cerevisiae'' (Kuge & Jones, 1994 [http://www.ncbi.nlm.nih.gov/pubmed?term=8313910], Shore & Nasmyth, 1987 [http://www.ncbi.nlm.nih.gov/pubmed?term=3315231], Schueller et al., 1994 [http://www.ncbi.nlm.nih.gov/pubmed?term=7523111], Ruis & Schueller, 1995 [http://www.ncbi.nlm.nih.gov/pubmed?term=8526890]). | ||
</div> | </div> | ||
| - | == General Remarks | + | == General Remarks == |
---- | ---- | ||
<div> | <div> | ||
| - | + | ||
| + | ''S. cerevisiae'' is a very potent brewer and quickly produces alcohol. This made negative controls during the characterization of the KlADH4-promoter difficult. | ||
</div> | </div> | ||
| + | |||
== Results == | == Results == | ||
---- | ---- | ||
| - | === | + | ===Extraction of the KlADH4-promoter from genomic DNA of ''K. lactis'' === |
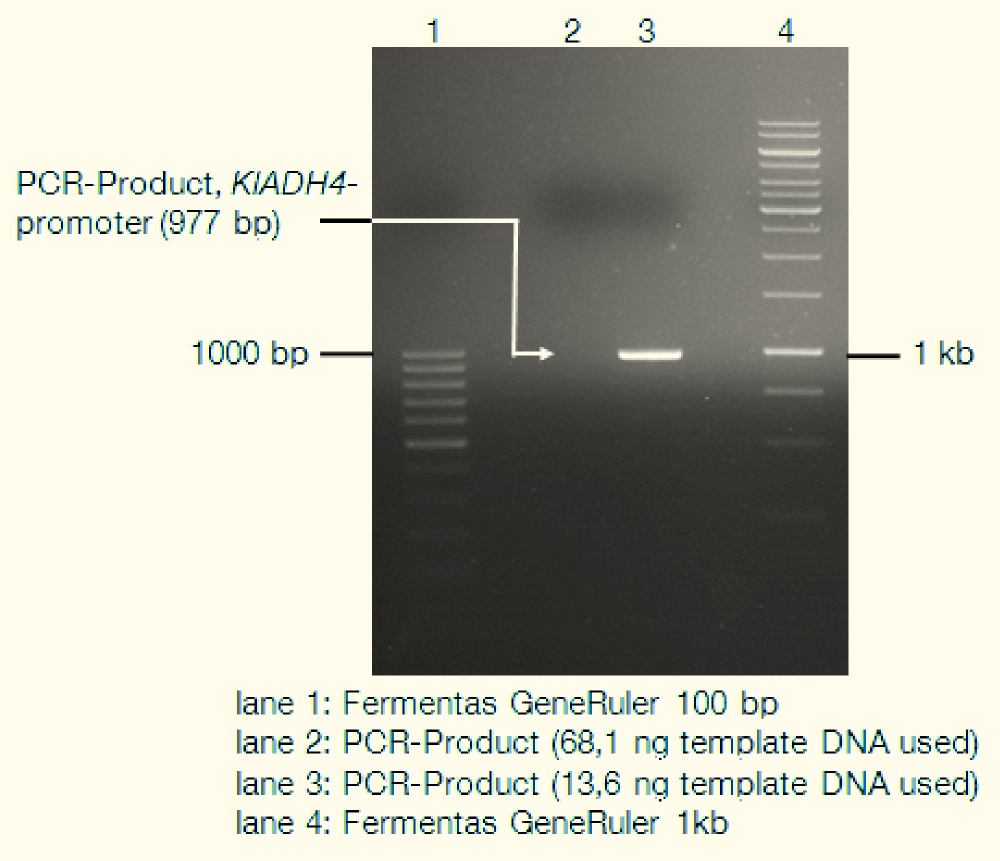
| - | <div> | + | <div>[[File:TUM12_KlADH4_gel.png|thumb|right|400px|lane 1: Fermentas GeneRuler 100bp; lane 2: empty; lane 3: PCR-Product (KlADH4-promoter); lane 4: Fermentas GeneRuler 1kb]] |
| + | We isolated genomic DNA from ''K. lactis'' using the LiOAc-SDS-method ([http://www.ncbi.nlm.nih.gov/pubmed?term=21548894 Looke et al. 2011]) and used this gDNA as template for PCR. The following gel picture documents the success of this: | ||
| + | |||
| + | |||
| + | |||
| + | We also sequenced the construct for verification (data not shown) and found some single point mutations compared to the published genome sequence of ''K. lactis''. These are not in any conserved regions and should have no influence on the promoters functionality. | ||
| + | The part was entered into the Partsregistry as [[http://partsregistry.org/Part:BBa_K801020 BBa_K801020]]. | ||
</div> | </div> | ||
| - | === | + | <hr> |
| + | |||
| + | === Characterization of the KlADH4-promoter using eGFP === | ||
<div> | <div> | ||
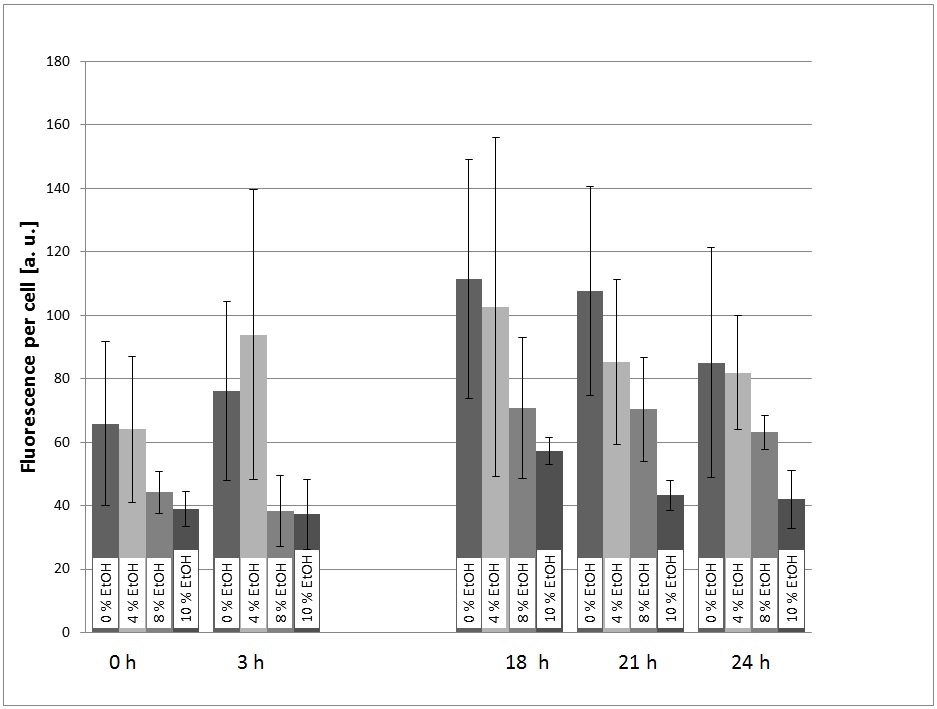
| + | ====First experiment using over-night pre-cultures==== | ||
| + | [[Image:TUM12_KlADH4_eGFP-expression_Graph1.jpg|400px|thumb|right|Figure 1: First characterization experiment of the KlADH4-promoter (& eGFP) in ''S. cerevisiae''. After an overnight pre culture, the transformed yeast cells were transferred into SC-U Media with different ethanol concentration and the eGFP-fluorescence and the OD600 were measured at different times.]] | ||
| + | In a first experiment, the transformed yeast cells were picked grown in a pre-culture (SC-U Medium, 30 °C, 180 rpm) over night and transferred into SC-U Medium with different concentrations of ethanol (0%, 4%, 8%, 10%, v/v). The eGFP-fluorescence and the OD600 were measured at t = 0h, 3h, 18h, 21h, 24h. Also, the ethanol concentration of the cultures was measured using an Alchohol-Dehydrogenase-Assay. | ||
| + | |||
| + | For the evaluation of the experimental data, the measured fluorescence was divided by the respecitve OD600, to normalize the fluorescence to the cell count. This was done to take the intrinsic auto-fluorescence in account. The results are shown in picture 1. | ||
| + | |||
| + | The promoter is generally functional in ''S. cerevisiae'', which can be seen by the fact that eGFP is expressed. | ||
| + | |||
| + | The fact that the expression of eGFP is low in the cultures with 8% and 10% (v/v) ethanol can be explained with the fact that the viability of the yeast cells is dramatically decreased at these high ethanol concentrations. | ||
| + | |||
| + | At first glance, the fact that there is a significant signal in the culture with 0% ethanol added looks as if the promoter is constitutive and not specifically induced by ethanol. However, the cells could also be induced by the ethanol produced by the yeast themselves during the over night pre-culture. Because of this ambiguity, further experiments were performed. | ||
| + | |||
| + | <hr> | ||
| + | |||
| + | ====Second experiment: Characterization of the KlADH4-promoter without over-night pre-cultures==== | ||
| + | |||
| + | The goal of this series of experiments was to avoid pre-cultures in which the cells would produce alcohol and hence be induced before the data was collected. Therefore, three measures were taken: | ||
| + | |||
| + | * Use of baffled flasks to increas oxygen transfer into the medium, thus lowering fermentation | ||
| + | * Transfer of a large amount of transformed yeast cells into SC-U Medium with different ethanol concentrations immediately after transformation and start measuring the eGFP-expression. The transformed cells were not plated on selective plates and thus, picking of clones and a pre-culture became unneccesary. (Experiment 2.1) | ||
| + | * Cultivate a control culture of transformed yeast in SC-U Medium with Glycerol as sole carbon source. Glycerol is a non-fermentable carbon source for ''S. cerevisiae'' (Feldmann, 2005). (Experiment 2.2) | ||
| + | |||
| + | '''Result of experiment 2.1: cells transferred into liquid medium immediately after transformation''' | ||
| + | |||
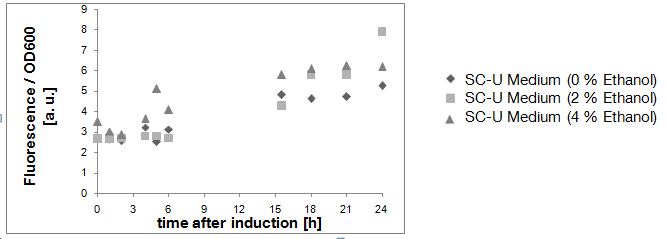
| + | [[Image:TUM12_KlADH4_after_transformation.png|800px|thumb|center|Figure 2: Fluorescence per cell of yeast cells transformed with a plasmid carrying BBa_K801020 in front of an eGFP-gene. The different alcohol concentrations are indicated]] | ||
| + | |||
| + | Due to the fact that the cultures examined here also contain untransformed yeast cells, the OD600-value is falsely high. The untransformed yeast cells are not viable (due to uracil auxotrophy and cultivation in SC-U, a medium lacking uracil) and cannot express eGFP, but still they disperse light and thus increase the measured OD. This renders the data shown in Fig. 2 uninterpretable. However, the cultures which were used for these measurements expressed eGFP - enough to produce fluorescence visible to the naked eye (Fig. 3). | ||
| + | |||
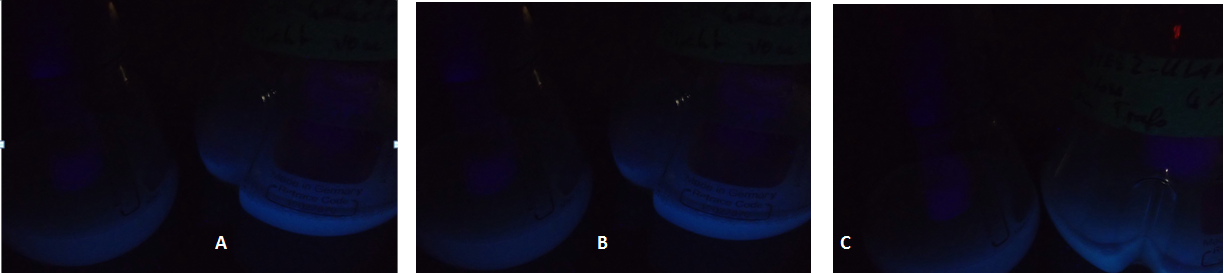
| + | [[Image:TUM12_KlADH4_fluorescent_cultures.png|800px|thumb|center|Figure 3: Fluorescent yeast cells carrying a plasmid with the KlADH4-promoter controlling eGFP expression. In each picture, the flask to the left shows a culture in which eGFP-expression is repressed by using the GAL1-Promoter and a medium containing glucose. A: Culture 1: 0 % Ethanol added at t = 0. B: Culture 2: 2 % Ethanol added at t = 0. C: Culture 3: 4 % Ethanol added at t = 0]] | ||
| + | |||
| + | '''Result of experiment 2.2: negative culture grown in SC-U glycerol''' | ||
| + | |||
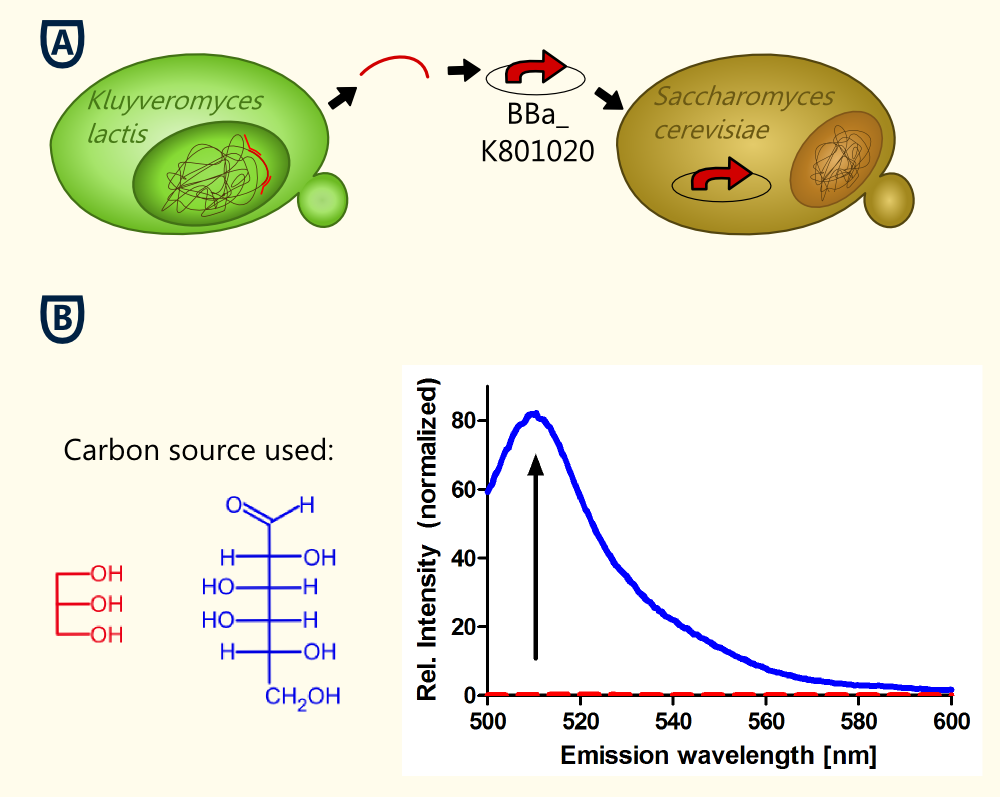
| + | [[file:TUM12_experiment_overwiew_alcohol1.png|400px|thumb|right| Figure 4: '''A''': The KlADH4-promoter was cloned from genomic DNA of ''Kluyveromyces lactis''. The new BioBrick BBa_K801020 was inserted into our pTUM100 vector. eGFP served as a reporter gene for characterization in ''S. cerevisiae'' (plasmid name: pTUM100_KLADH4_eGFP). '''B''' Emission spectra of eGFP obtained during cultivation of ''S. cerevisiae'' transformed with pTUM100_KLADH4_eGFP using different carbon sources. Blue: Galactose was used as carbon source. The measured ethanol concentration was 1.7 % (v/v). The peak at 509 nm indicates that eGFP is expressed. Red: Glycerol was used as carbon source. The measured ethanol concentration was 0.2 % (v/v). No eGFP fluorescence could be detected.]] | ||
| + | |||
| + | The aim of this experiment was to keep the ethanol production as low as possible by the use of glycerol as sole carbon source. The ethanol concentration in the culture remained very low (less that 0.21 Vol.-%). Also, the "Fluorescence per cell" did not increase over time. | ||
| + | |||
| + | To verify the fact that the cells did not express eGFP, an emission spectrum was recorded at t = 24 h. For comparison, a reference spectrum of a culture expressing eGFP is also shown (Fig. 4). The characteristic peak of eGFP (emission: 509 nm) is not visible in the culture grown on glycerol as sole carbon source (ethanol concentration: 0,2 Vol.-%). | ||
| + | |||
| + | The fact that no expression of eGFP is detected in this culture (ethanol concentration: 0.2 Vol.-%) '''suggests that the part is ethanol inducible in ''S. cerevisiae'', too'''. The lowest measured alcohol concentration in a culture expressing eGFP was 0.9 Vol.-%. Further experiments are being done to verify this first statement. | ||
</div> | </div> | ||
| Line 64: | Line 115: | ||
---- | ---- | ||
* Breunig KD., Bolotin-Fukuhara M., Bianchi MM., Bourgarel D., Falcone C., Ferrero I I., Frontali L., Goffrini P., Krijger JJ., Mazzoni C., Milkowski C., Steensma HY., Wésolowski-Louvel M., Zeeman AM. (2000),'Regulation of primary carbon metabolism in Kluyveromyces lactis', ''Enzyme Microb Technol.'' '''26 (9-10)''', 771-780. PMID: 10862884 | * Breunig KD., Bolotin-Fukuhara M., Bianchi MM., Bourgarel D., Falcone C., Ferrero I I., Frontali L., Goffrini P., Krijger JJ., Mazzoni C., Milkowski C., Steensma HY., Wésolowski-Louvel M., Zeeman AM. (2000),'Regulation of primary carbon metabolism in Kluyveromyces lactis', ''Enzyme Microb Technol.'' '''26 (9-10)''', 771-780. PMID: 10862884 | ||
| + | |||
| + | * Studier, FW. (2005), Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 41:207-34. PMID: 15915565 | ||
* Mazzoni, C., Santori, F., Saliola, M. & Falcone, C. (2000) ‚Molecular analysis of UASE, a cis element containing stress response elements responsible for ethanol induction of the KlADH4 gene of Kluyveromyces lactis’, ''Res. Microbiol.'' '''151''', 19-28. PMID: 10724480 | * Mazzoni, C., Santori, F., Saliola, M. & Falcone, C. (2000) ‚Molecular analysis of UASE, a cis element containing stress response elements responsible for ethanol induction of the KlADH4 gene of Kluyveromyces lactis’, ''Res. Microbiol.'' '''151''', 19-28. PMID: 10724480 | ||
* Kuge S., Jones N. (1994) ,YAP1 dependent activation of TRX2 is essential for the response of ''S. cerevisiae'' to oxidative stress by hydroperoxides', ''EMBO J.'' '''13''', 655–664. PMID: 8313910 | * Kuge S., Jones N. (1994) ,YAP1 dependent activation of TRX2 is essential for the response of ''S. cerevisiae'' to oxidative stress by hydroperoxides', ''EMBO J.'' '''13''', 655–664. PMID: 8313910 | ||
| + | |||
| + | * Saliola, M., Mazzoni, C., Solimando, N., Crisà, A.,Falcone, C. & Jung, G. (1999) ‚Use of the KlADH4 promoter for ethanol-dependent production of recombinant human serum albumin in Kluyveromyces lactis’, Appl Environ Microbiol. 65(1), 53-60 | ||
* Shore D., Nasmyth K. (1987) ,Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements', ''Cell'' '''51 (5)''' 721-32. PMID: 3315231 | * Shore D., Nasmyth K. (1987) ,Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements', ''Cell'' '''51 (5)''' 721-32. PMID: 3315231 | ||
Latest revision as of 22:31, 26 October 2012



Contents |
Ethanol Inducible Promoter
An ethanol inducible promoter - that would really be handy in brewing a beer with transgenic yeast! According to the German Purity Law, no ingredients other than water, hops and barley may be used for brewing - which is why we can't simply add a chemical to induce gene expression at the desired point of time. But constitutive promoters are not always the best solution, so we thought about substances that "appear" during brewing. And of course we came up with - Alcohol! Alcohol would be the ideal inducing agent because the yeast cells could take care of the main fermentation first, without any heterologous pathways hindering them. Then, after the main fermentation is completed, ethanol is present and induce the heterologous genes - the yeast produces our desired substances.
Also beyond the scope of brewing an ethanol sensitive promoter would have a broad spectrum of application. If the induction of heterologous gene expression occurs at sufficiently high ethanol concentrations, a system similar to the T7-promoter based auto-induction system in E. coli (Studier, 2005) could be established.
Another application for an alcohol inducible promoter is that of a classic biosensor. If cloned in front of a reporter gene such as GFP, the fluorescence could be used to deduce the current ethanol content, thus abolishing the need for complicated enzyme assays or distillation experiments, which are currently among the most applied tools for measuring the alcohol content in beverages.
We chose the KlADH4-promoter because it originates from a close relative of S. cerevisiae - Kluyveromyces lactis. It has been reported that promoters can be interchanged between those two species and in addition to that, all transcription factors and cis-elemtents known to be involved in the alcohol response of the KlADH4 promoter in K. lactis also occur in S. cerevisiae (Mazzoni et al., 2000). Because of that, we do not need to express genes for any additional transcription factors.
Background and principles
Original organism: Kluyveromyces lactis
K. lactis is a crabtree-negative yeast, which means it does not produce ethanol under aerobic conditions. S. cerevisiae, on the other hand, is crabtree-positive. Another interesting fact about K. lactis is its ability to grow on ethanol as the only carbon source (Breunig et al., 1999, [http://www.ncbi.nlm.nih.gov/pubmed?term=10862884]).
KlADH4
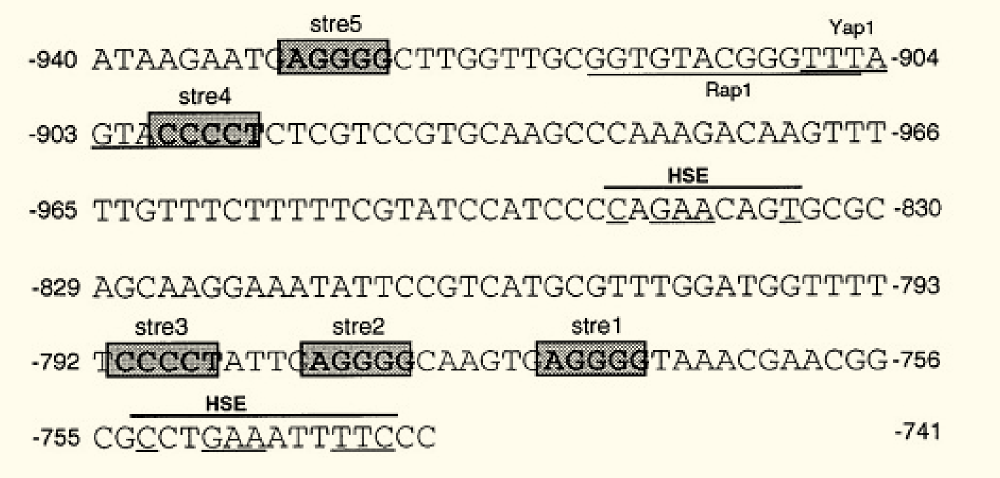
The K. lactis gene KlADH4 encodes a mitochondrial alcohol dehydrogenase which can be specifically induced by ethanol and is insensitive to glucose repression. A small region in the KlADH4 promoter, called UASE, renders the expression of a gene ethanol-dependent (Mazzoni et al., 2000 [http://www.ncbi.nlm.nih.gov/pubmed?term=10724480]). UASE is a cis-element of the KlADH4 promoter spanning from -953 to -741. When inserted into the promoters of heterologous genes, these genes become ethanol-inducible in K lactis.
The sequence contains (Mazzoni et al. 2000):
- a putative binding site for Yap1p between -906/-900, a transcription factor involved in stress response in S. cerevisiae and K. lactis
- consensus sequence of the binding site of Rap1 protein. RAP1 (repressor activator protein 1) is an essential gene present in both S. cerevisiae and K. lactis and acts positively or negatively on the expression of many genes. Rap1 binding sites have been found in promoters of glycolytic genes where they play a positive role in transcription.
- Five STREs (stress response elements). STREs have been found in the promoters of many S. cerevisiae genes, where they are present in two or more copies in both orientations
- Two HSEs (heat shock elements). HSEs have been found in combination with STREs in many stress-responsive genes of S. cerevisiae.
Despite the presence of the STREs, KlADH4 is not a stress responsive gene but specifically induced by alcohol (Mazzoni et al., 2000).
Idea
Although there is evidence that the KlADH4 promoter can be used for ethanol dependent production of recombinant proteins in K. lactis ([http://www.ncbi.nlm.nih.gov/pubmed/9872759 Saliola et al. 1999]), we did not find any literature which indicated that this alcohol-inducible promoter has been used in other organisms such as S. cerevisiae.
Mazzoni et. al (2000) annotated binding sites for two transcription factors (Rap1 and Yap1) as well as some small cis-elements (HSEs and STREs) in the UASE-region of the KlADH4-promoter. All of these factors are not only present in K. lactis but also in S. cerevisiae (Kuge & Jones, 1994 [http://www.ncbi.nlm.nih.gov/pubmed?term=8313910], Shore & Nasmyth, 1987 [http://www.ncbi.nlm.nih.gov/pubmed?term=3315231], Schueller et al., 1994 [http://www.ncbi.nlm.nih.gov/pubmed?term=7523111], Ruis & Schueller, 1995 [http://www.ncbi.nlm.nih.gov/pubmed?term=8526890]).
General Remarks
S. cerevisiae is a very potent brewer and quickly produces alcohol. This made negative controls during the characterization of the KlADH4-promoter difficult.
Results
Extraction of the KlADH4-promoter from genomic DNA of K. lactis
We isolated genomic DNA from K. lactis using the LiOAc-SDS-method ([http://www.ncbi.nlm.nih.gov/pubmed?term=21548894 Looke et al. 2011]) and used this gDNA as template for PCR. The following gel picture documents the success of this:
We also sequenced the construct for verification (data not shown) and found some single point mutations compared to the published genome sequence of K. lactis. These are not in any conserved regions and should have no influence on the promoters functionality.
The part was entered into the Partsregistry as http://partsregistry.org/Part:BBa_K801020 BBa_K801020.
Characterization of the KlADH4-promoter using eGFP
First experiment using over-night pre-cultures
In a first experiment, the transformed yeast cells were picked grown in a pre-culture (SC-U Medium, 30 °C, 180 rpm) over night and transferred into SC-U Medium with different concentrations of ethanol (0%, 4%, 8%, 10%, v/v). The eGFP-fluorescence and the OD600 were measured at t = 0h, 3h, 18h, 21h, 24h. Also, the ethanol concentration of the cultures was measured using an Alchohol-Dehydrogenase-Assay.
For the evaluation of the experimental data, the measured fluorescence was divided by the respecitve OD600, to normalize the fluorescence to the cell count. This was done to take the intrinsic auto-fluorescence in account. The results are shown in picture 1.
The promoter is generally functional in S. cerevisiae, which can be seen by the fact that eGFP is expressed.
The fact that the expression of eGFP is low in the cultures with 8% and 10% (v/v) ethanol can be explained with the fact that the viability of the yeast cells is dramatically decreased at these high ethanol concentrations.
At first glance, the fact that there is a significant signal in the culture with 0% ethanol added looks as if the promoter is constitutive and not specifically induced by ethanol. However, the cells could also be induced by the ethanol produced by the yeast themselves during the over night pre-culture. Because of this ambiguity, further experiments were performed.
Second experiment: Characterization of the KlADH4-promoter without over-night pre-cultures
The goal of this series of experiments was to avoid pre-cultures in which the cells would produce alcohol and hence be induced before the data was collected. Therefore, three measures were taken:
- Use of baffled flasks to increas oxygen transfer into the medium, thus lowering fermentation
- Transfer of a large amount of transformed yeast cells into SC-U Medium with different ethanol concentrations immediately after transformation and start measuring the eGFP-expression. The transformed cells were not plated on selective plates and thus, picking of clones and a pre-culture became unneccesary. (Experiment 2.1)
- Cultivate a control culture of transformed yeast in SC-U Medium with Glycerol as sole carbon source. Glycerol is a non-fermentable carbon source for S. cerevisiae (Feldmann, 2005). (Experiment 2.2)
Result of experiment 2.1: cells transferred into liquid medium immediately after transformation
Due to the fact that the cultures examined here also contain untransformed yeast cells, the OD600-value is falsely high. The untransformed yeast cells are not viable (due to uracil auxotrophy and cultivation in SC-U, a medium lacking uracil) and cannot express eGFP, but still they disperse light and thus increase the measured OD. This renders the data shown in Fig. 2 uninterpretable. However, the cultures which were used for these measurements expressed eGFP - enough to produce fluorescence visible to the naked eye (Fig. 3).
Result of experiment 2.2: negative culture grown in SC-U glycerol
The aim of this experiment was to keep the ethanol production as low as possible by the use of glycerol as sole carbon source. The ethanol concentration in the culture remained very low (less that 0.21 Vol.-%). Also, the "Fluorescence per cell" did not increase over time.
To verify the fact that the cells did not express eGFP, an emission spectrum was recorded at t = 24 h. For comparison, a reference spectrum of a culture expressing eGFP is also shown (Fig. 4). The characteristic peak of eGFP (emission: 509 nm) is not visible in the culture grown on glycerol as sole carbon source (ethanol concentration: 0,2 Vol.-%).
The fact that no expression of eGFP is detected in this culture (ethanol concentration: 0.2 Vol.-%) suggests that the part is ethanol inducible in S. cerevisiae, too. The lowest measured alcohol concentration in a culture expressing eGFP was 0.9 Vol.-%. Further experiments are being done to verify this first statement.
References
- Breunig KD., Bolotin-Fukuhara M., Bianchi MM., Bourgarel D., Falcone C., Ferrero I I., Frontali L., Goffrini P., Krijger JJ., Mazzoni C., Milkowski C., Steensma HY., Wésolowski-Louvel M., Zeeman AM. (2000),'Regulation of primary carbon metabolism in Kluyveromyces lactis', Enzyme Microb Technol. 26 (9-10), 771-780. PMID: 10862884
- Studier, FW. (2005), Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 41:207-34. PMID: 15915565
- Mazzoni, C., Santori, F., Saliola, M. & Falcone, C. (2000) ‚Molecular analysis of UASE, a cis element containing stress response elements responsible for ethanol induction of the KlADH4 gene of Kluyveromyces lactis’, Res. Microbiol. 151, 19-28. PMID: 10724480
- Kuge S., Jones N. (1994) ,YAP1 dependent activation of TRX2 is essential for the response of S. cerevisiae to oxidative stress by hydroperoxides', EMBO J. 13, 655–664. PMID: 8313910
- Saliola, M., Mazzoni, C., Solimando, N., Crisà, A.,Falcone, C. & Jung, G. (1999) ‚Use of the KlADH4 promoter for ethanol-dependent production of recombinant human serum albumin in Kluyveromyces lactis’, Appl Environ Microbiol. 65(1), 53-60
- Shore D., Nasmyth K. (1987) ,Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements', Cell 51 (5) 721-32. PMID: 3315231
- Schüller C, Brewster JL, Alexander MR, Gustin MC, Ruis H. (1994) ,The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene', EMBO J. 13(18) 4382-9. PMID: 7523111
- Ruis H, Schüller C. (1995) ,Stress signaling in yeast',Bioessays. 17(11) 959-65. PMID: 8526890
 "
"