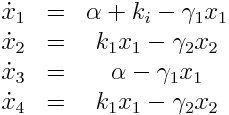Team:TU Munich/Modeling/Gal1 Promoter
From 2012.igem.org
(→Joint Distribution) |
(→Joint Distribution) |
||
| Line 97: | Line 97: | ||
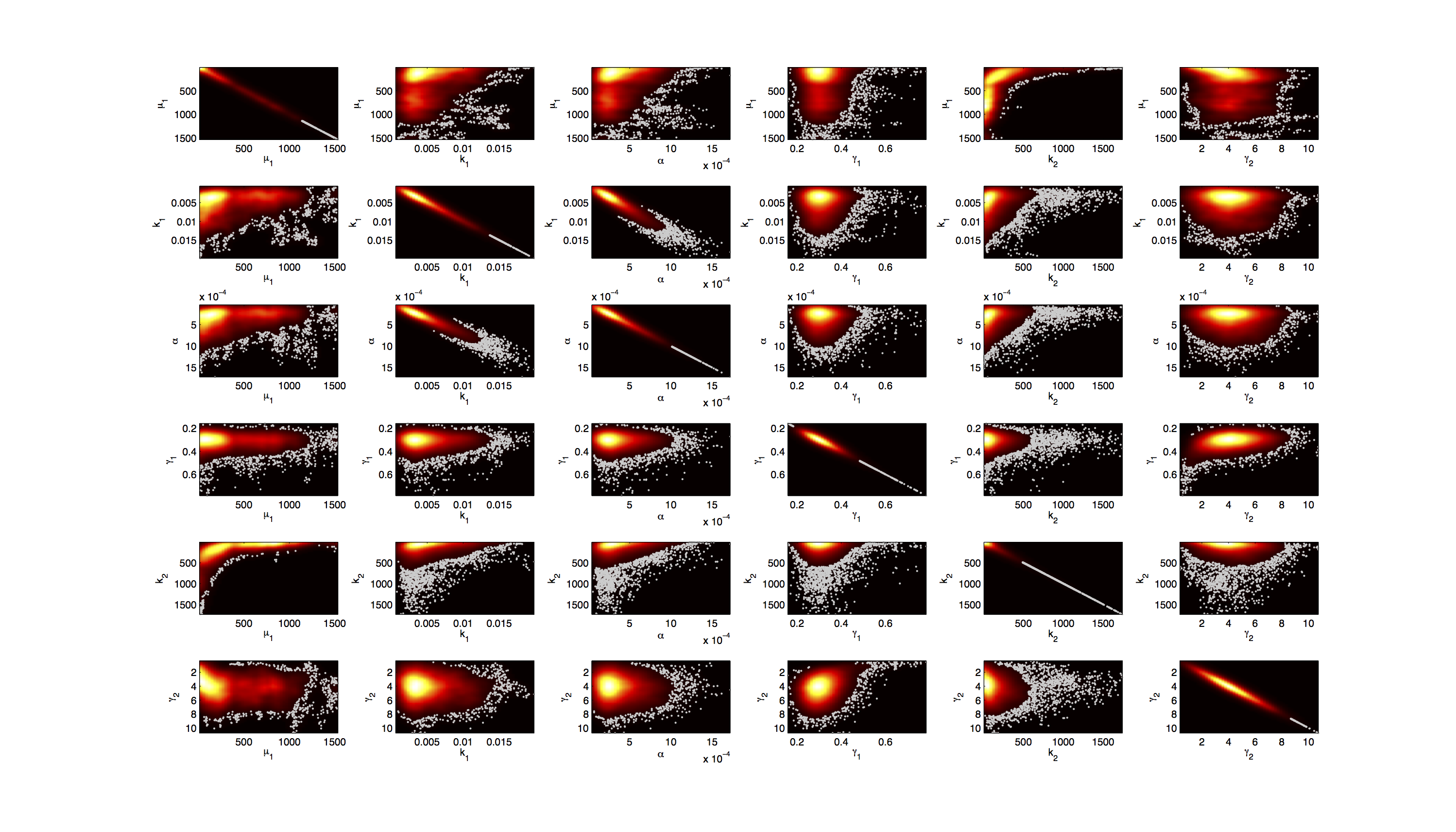
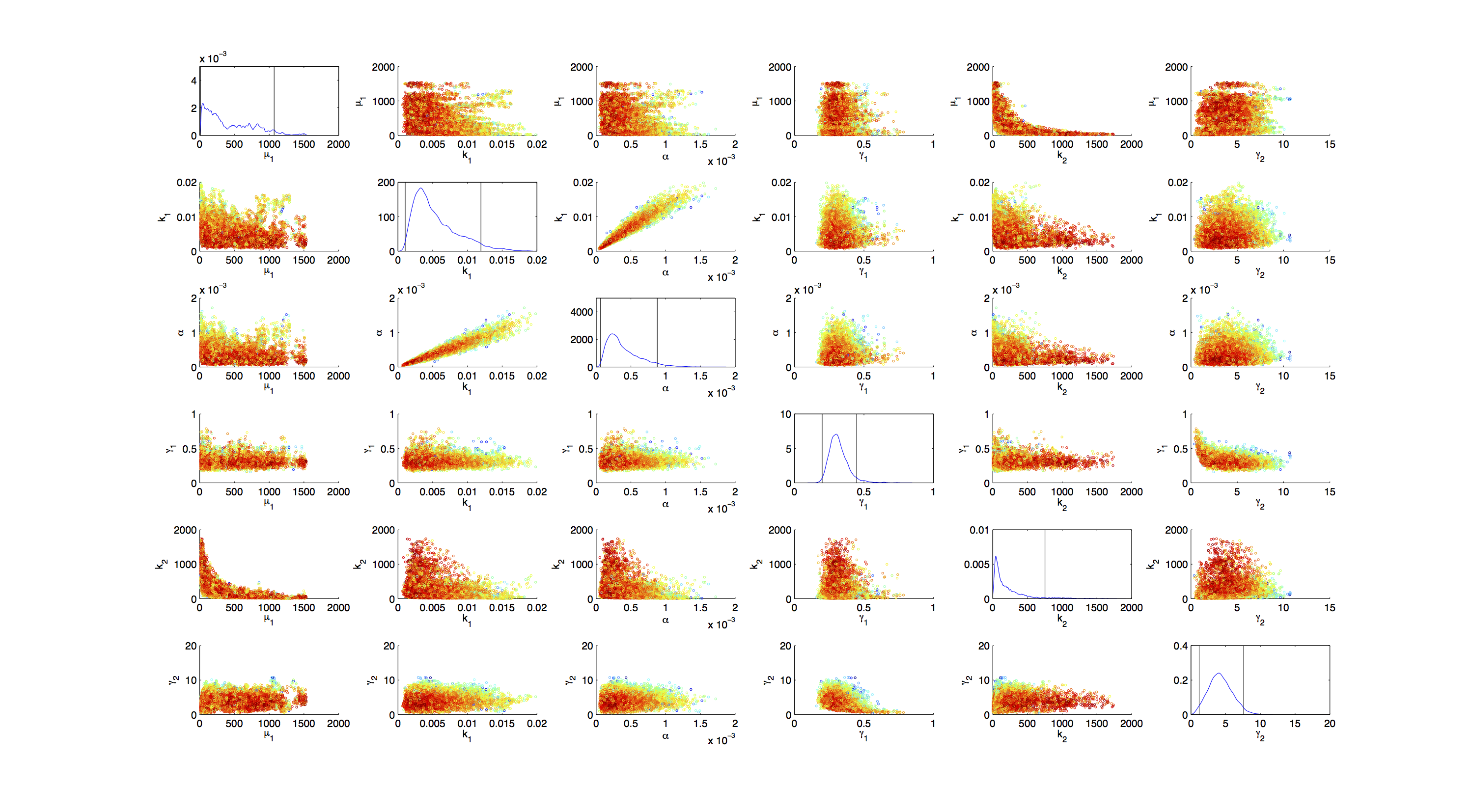
[[File:TUM12_GAL1_Scatter.png|400px|thumb|right|Figure 3. Scatter plot of the joint distribution of in each case 2 parameters.]] | [[File:TUM12_GAL1_Scatter.png|400px|thumb|right|Figure 3. Scatter plot of the joint distribution of in each case 2 parameters.]] | ||
| + | <div style="clear:both"> | ||
For the Scatter plot further thinning of 1:1000 was necessary for scatter.m to be able to process the data. | For the Scatter plot further thinning of 1:1000 was necessary for scatter.m to be able to process the data. | ||
On the diagonal the kernel density estimation script [http://www.mathworks.com/matlabcentral/fileexchange/14034-kernel-density-estimator kde.m] was used to obtain a plot of the density. | On the diagonal the kernel density estimation script [http://www.mathworks.com/matlabcentral/fileexchange/14034-kernel-density-estimator kde.m] was used to obtain a plot of the density. | ||
| + | </div> | ||
<div style="clear:both"> | <div style="clear:both"> | ||
Revision as of 13:51, 16 September 2012



Contents |
Gal1 Promoter
The Gal1 Promoter is the standard promoter for our pYES vector that is used in the expression of all ingredient pathways. As it is also part of the construct for the light switchable promoter, it is crucial to thoroughly characterize the kinetics of this promoter.
Model Specification
Model Equations
We used a two species approach for mRNA and Protein concentrations. x1 and x2 represent the mRNA and protein levels for the induced promoter and x3 and x4 the respective levels for the uninduced promoter.
As no analysis was done with several galactose concentrations hence the hill function that normally models the response to the concentration was replaced by a single factor to improve identifiability.
Parameters
| Name | Description | Prior? | Best fit | Unit |
| μ | Scaling factor | NO | 114.9 | - |
| ki | Induced transcription rate | YES | 0.006758 | mol/h |
| α | Leaky transcription rate | NO | 0.0004784 | mol/h |
| γ1 | mRNA degradation rate | YES | 0.2892 | 1/h |
| k2 | Protein synthesis rate | NO | 165.9 | mol/h |
| γ2 | Protein degradation rate | NO | 3.1874 | 1/h |
| σ1 | Standard deviation for measured data of induced cells | NO | 19.55 | - |
| σ2 | Standard deviation for measured data of noninduced cells | NO | 5.337 | - |
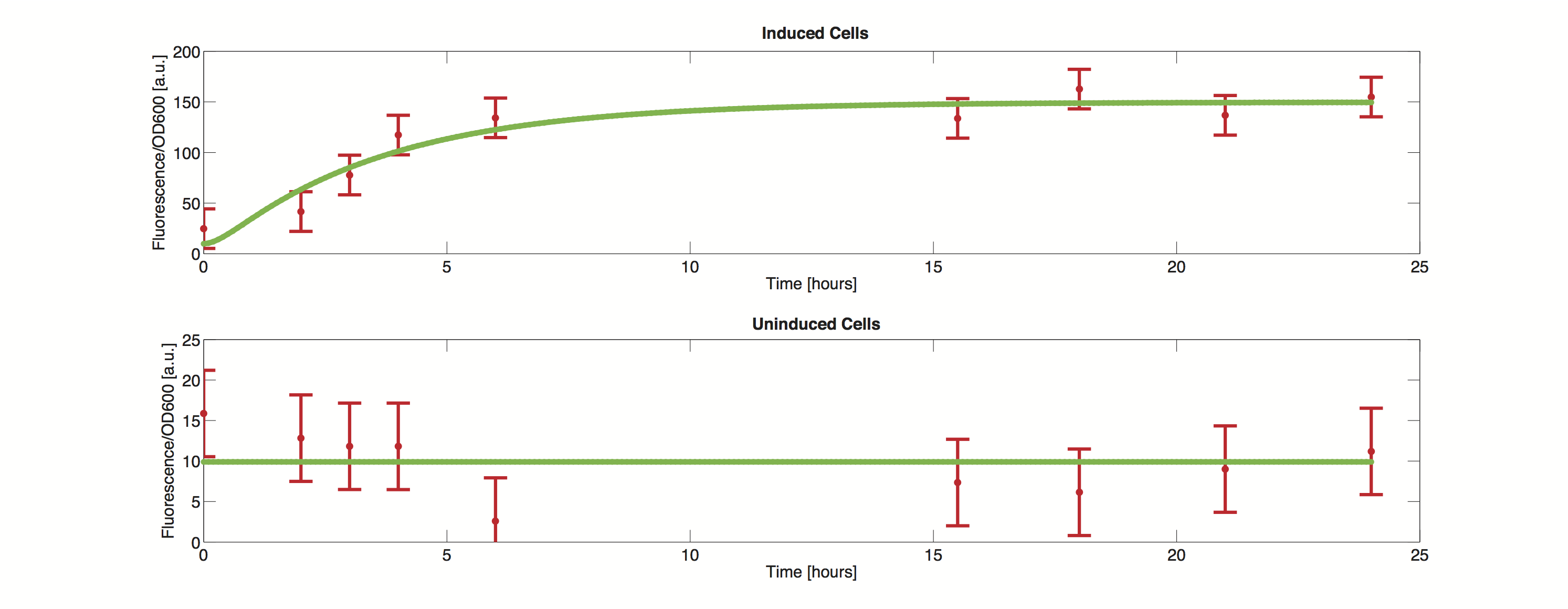
Data
The methods utilized rely on a value for standard deviation, but only one experimental measurement was performed. Hence a value was infered during the optimization process. Thus the error bars in the plot do not reflect values from actual measurements but merely show how well the data can be approximated by the model.
Profile Likelihood
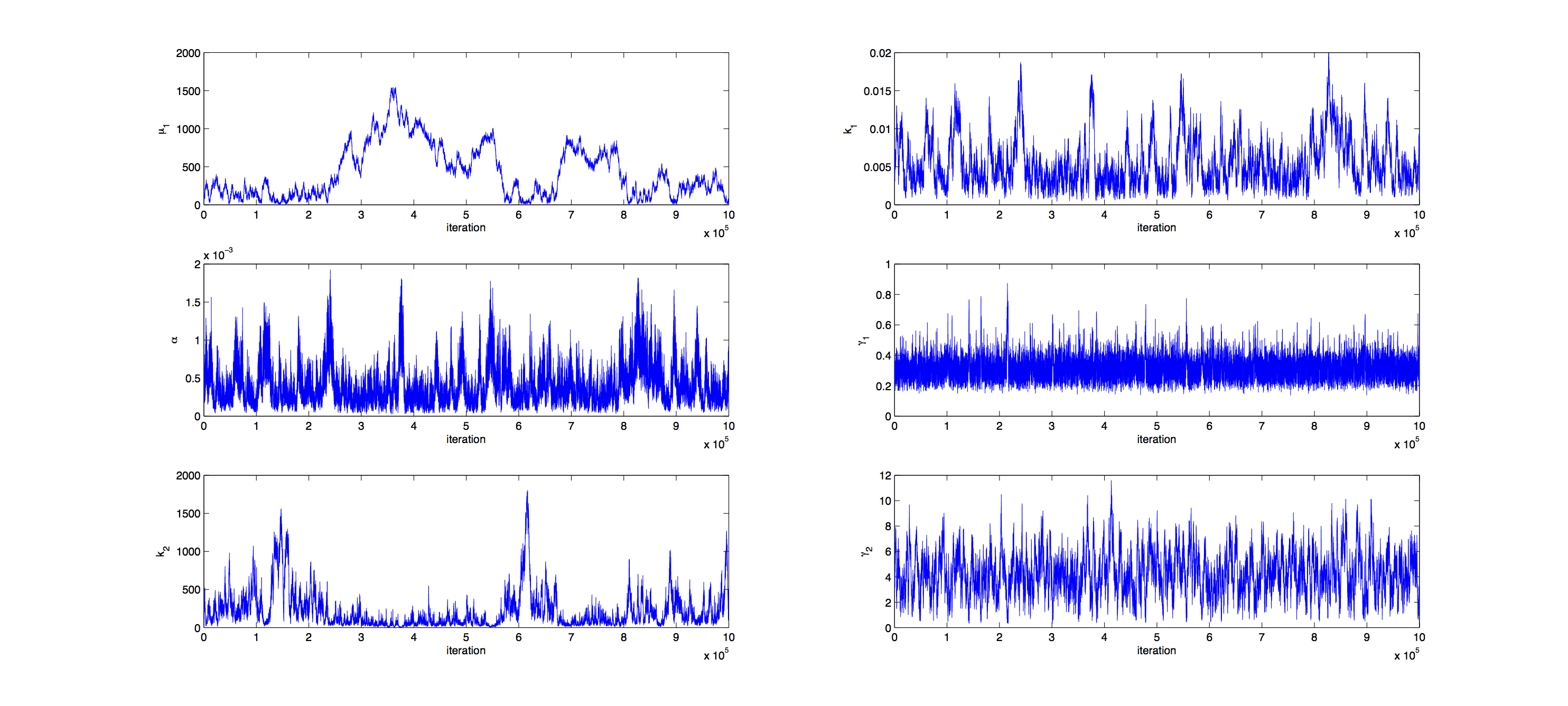
Markov Analysis
Analysis was performed with a Metropolis Hastings algorithm. 1000000 samples were generated and then thinned by a factor 1:100 to reduce correlation of the samples. The acceptance rate for the samples was 16% so a little below the target of 23%.
Joint Distribution
For the Scatter plot further thinning of 1:1000 was necessary for scatter.m to be able to process the data. On the diagonal the kernel density estimation script kde.m was used to obtain a plot of the density.
Credibility Intervals
As the Metropolis Hastings algorithm samples exactly from the target distribution, one can use the generated samples to find credibility intervals. The shown intervals are maximum density credibility intervals. They were obtained by computing the 1-α/2; and α/2 quantiles, with alpha being the missing percentage to 100%, that minimized the 1-norm of the distance between the two values.
95% Credibility Interval for μ1: [7.6525 , 203.2962]
95% Credibility Interval for k1: [0.0011078 , 0.017724]
95% Credibility Interval for α: [8.0055e-05 , 0.001293]
95% Credibility Interval for γ1: [0.20397 , 0.45586]
95% Credibility Interval for k2: [24.3219 , 954.8224]
95% Credibility Interval for γ2: [1.0073 , 7.3142]
75% Credibility Interval for μ1: [14.3725 , 116.5086]
75% Credibility Interval for k1: [0.0020553 , 0.0089666]
75% Credibility Interval for α: [0.00012838 , 0.00064765]
75% Credibility Interval for γ1: [0.24028 , 0.3761]
75% Credibility Interval for k2: [126.3764 , 646.5652]
75% Credibility Interval for γ2: [1.8417 , 5.5854]
50% Credibility Interval for μ1: [14.7945 , 73.2214]
50% Credibility Interval for k1: [0.002091 , 0.0058511]
50% Credibility Interval for α: [0.00017481 , 0.00044449]
50% Credibility Interval for γ1: [0.25771 , 0.33486]
50% Credibility Interval for k2: [327.0597 , 639.2103]
50% Credibility Interval for γ2: [2.4019 , 4.6641]
Sensitivity Analysis
== Reference ==
 "
"