Team:TU Darmstadt/Labjournal/Metabolism
From 2012.igem.org
(Difference between revisions)
| Line 48: | Line 48: | ||
* Reconstitution of ''C. testosteroni KF-1'' according to DSMZ [http://www.dsmz.de/fileadmin/Bereiche/Microbiology/Dateien/Kultivierungshinweise/engl_Opening.pdf protocol] | * Reconstitution of ''C. testosteroni KF-1'' according to DSMZ [http://www.dsmz.de/fileadmin/Bereiche/Microbiology/Dateien/Kultivierungshinweise/engl_Opening.pdf protocol] | ||
* [[Cultivation]] of ''C. testosteroni KF-1'' on agar plates with [[Medium 1]] | * [[Cultivation]] of ''C. testosteroni KF-1'' on agar plates with [[Medium 1]] | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Production_of_chemically_competent_cells Production of chemically ] competent ''E. coli DH5α'' and ''E. coli BL21(DE3)pLysS'' cells |
==week 2 (21.-25.05.12)== | ==week 2 (21.-25.05.12)== | ||
'''tphA1''' | '''tphA1''' | ||
| - | * Isolation of the gene from ''C. testosteroni KF-1'' genome using [ | + | * Isolation of the gene from ''C. testosteroni KF-1'' genome using [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] |
** Annealing temperature: 49 °C | ** Annealing temperature: 49 °C | ||
** [[Primer]]: tphA1-l-F and tphA1-l-R | ** [[Primer]]: tphA1-l-F and tphA1-l-R | ||
| - | ** Both PCR products were purified via [ | + | ** Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | ** Concentrations measured by [ | + | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
::{| class="wikitable" | ::{| class="wikitable" | ||
|- | |- | ||
| Line 65: | Line 65: | ||
'''tphA3''' | '''tphA3''' | ||
| - | * Isolation of the gene from ''C. testosteroni KF-1'' genome using [ | + | * Isolation of the gene from ''C. testosteroni KF-1'' genome using [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] |
** Annealing temperature: 59 °C | ** Annealing temperature: 59 °C | ||
** [[Primer]]: tphA3-l-F and tphA3-l-R | ** [[Primer]]: tphA3-l-F and tphA3-l-R | ||
| - | ** Both PCR products were purified via [ | + | ** Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | ** Concentrations measured by [ | + | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
::{| class="wikitable" | ::{| class="wikitable" | ||
|- | |- | ||
| Line 78: | Line 78: | ||
'''tphB''' | '''tphB''' | ||
| - | * Isolation of the gene from ''C. testosteroni KF-1'' genome using [ | + | * Isolation of the gene from ''C. testosteroni KF-1'' genome using [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] |
** Annealing temperature: 59 °C | ** Annealing temperature: 59 °C | ||
** [[Primer]]: tphB-l-F and tphB-l-R | ** [[Primer]]: tphB-l-F and tphB-l-R | ||
| - | ** Both PCR products were purified via [ | + | ** Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | ** Concentrations measured by [ | + | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
::{| class="wikitable" | ::{| class="wikitable" | ||
|- | |- | ||
| Line 91: | Line 91: | ||
'''Other''' | '''Other''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] and [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Plasmid_Midiprep midi prep] of all used [[Biobricks]] |
| - | ** Concentrations measured by [ | + | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
::{| class="wikitable" | ::{| class="wikitable" | ||
|- | |- | ||
| Line 110: | Line 110: | ||
* Two PCRs were performed to mutate the PstI site in the wild type gene. The primer tphA1-l-PstI(99)-R and tphA1-l-PstI(99)-F respectively introduced a BsaI site | * Two PCRs were performed to mutate the PstI site in the wild type gene. The primer tphA1-l-PstI(99)-R and tphA1-l-PstI(99)-F respectively introduced a BsaI site | ||
** tphA1 fragment 1 | ** tphA1 fragment 1 | ||
| - | ** [ | + | ** [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR_on_a_DNA_template PCR] on tphA1 isolated from ''C. testosteroni'' |
*** Annealing temperature: 69 °C | *** Annealing temperature: 69 °C | ||
*** [[Primer]]: tphA1-l-PstI(99)-R and tphA1-l-R | *** [[Primer]]: tphA1-l-PstI(99)-R and tphA1-l-R | ||
** tphA1 fragment 2 | ** tphA1 fragment 2 | ||
| - | ** [ | + | ** [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR_on_a_DNA_template PCR] on tphA1 isolated from ''C. testosteroni'' |
*** Annealing temperature: 69 °C | *** Annealing temperature: 69 °C | ||
*** [[Primer]]: tphA1-l-PstI(99)-F and tphA1-l-F | *** [[Primer]]: tphA1-l-PstI(99)-F and tphA1-l-F | ||
| - | ** Both PCR products were purified via [ | + | ** Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | ** Concentrations measured by [ | + | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
::{| class="wikitable" | ::{| class="wikitable" | ||
|- | |- | ||
| Line 131: | Line 131: | ||
| - | * Both fragments were cut with BsaI in a [ | + | * Both fragments were cut with BsaI in a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] |
** The ligation mix differed from our standard protocol in the following manner | ** The ligation mix differed from our standard protocol in the following manner | ||
*** 100 ng of fragment 1 | *** 100 ng of fragment 1 | ||
| Line 139: | Line 139: | ||
*** add DI water up to 20 µL | *** add DI water up to 20 µL | ||
*** incubate for 15 minutes at 37 °C | *** incubate for 15 minutes at 37 °C | ||
| - | ** [ | + | ** [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR_on_a_DNA_template PCR] on ligation mix |
*** Annealing temperature: 59 °C | *** Annealing temperature: 59 °C | ||
*** [[Primer]]: tphA1-l-R and tphA1-l-F | *** [[Primer]]: tphA1-l-R and tphA1-l-F | ||
| - | ** The PCR product was purified via [ | + | ** The PCR product was purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | ** Concentrations measured by [ | + | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
::{| class="wikitable" | ::{| class="wikitable" | ||
|- | |- | ||
| Line 153: | Line 153: | ||
==week 4 (04.-08.06.12)== | ==week 4 (04.-08.06.12)== | ||
'''Other''' | '''Other''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] digest of BBa_K316003 by EcoRI and PstI |
| - | ** Purification of plasmid backbone pSB1C3 via [ | + | ** Purification of plasmid backbone pSB1C3 via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | ** Concentrations measured by [ | + | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
::{| class="wikitable" | ::{| class="wikitable" | ||
|- | |- | ||
| Line 163: | Line 163: | ||
|} | |} | ||
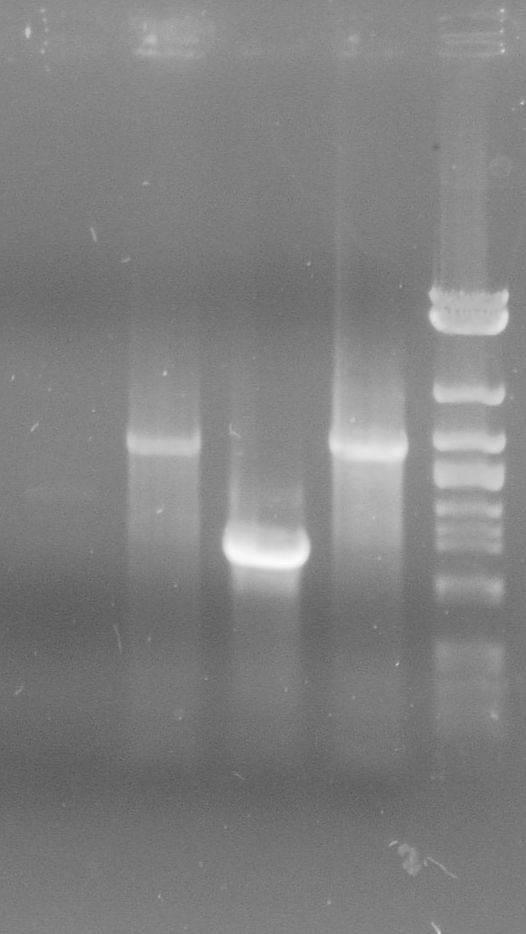
| - | [[File:WIKI-2012-06-06-_xylE_plasmid_restriction.jpg|thumb|none|alt=A| | + | [[File:WIKI-2012-06-06-_xylE_plasmid_restriction.jpg|thumb|none|alt=A|restriction of BBa_K316003 using EcoRI and PstI (1kb DNA ladder, NEB)]] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] digest of BBa_K316003 by XbaI and PstI |
| - | ** Purification of insert xylE-dT via [ | + | ** Purification of insert xylE-dT via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | ** Concentrations measured by [ | + | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
::{| class="wikitable" | ::{| class="wikitable" | ||
|- | |- | ||
| Line 178: | Line 178: | ||
'''Other''' | '''Other''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] digest of BBa_J23100 by SpeI and PstI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Dephosphorylation Dephosphorylation] of the restriction |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of BBa_J23100 (cut with SpeI and PstI) and xylE-dT (cut with XbaI and PstI) |
| - | ** [ | + | ** [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of the ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] of the transformation for verification |
* After overnight [[incubation]] of colony x in [[LB medium]] with ampicilin a [[glycerine stock]] was made | * After overnight [[incubation]] of colony x in [[LB medium]] with ampicilin a [[glycerine stock]] was made | ||
| Line 209: | Line 209: | ||
==week 10 (16.-20.07.12)== | ==week 10 (16.-20.07.12)== | ||
'''tphA1''' | '''tphA1''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR_on_a_DNA_template PCR] on mutated tphA1 |
** Annealing temperature: 59 °C | ** Annealing temperature: 59 °C | ||
** [[Primer]]: tphA1-Suffix_R and tphA1-l-Prefix | ** [[Primer]]: tphA1-Suffix_R and tphA1-l-Prefix | ||
| - | * Both PCR products were purified via [ | + | * Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 220: | Line 220: | ||
| Mutated tphA1-prefix/suffix || 62.0 | | Mutated tphA1-prefix/suffix || 62.0 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of mutated tphA1-prefix/suffix with EcoRI and PstI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of mutated tphA1-prefix/suffix (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_4_.2804.-08.06.12.29 pSB1C3 (cut with EcoRI and PstI)] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation |
** The PCR was negative | ** The PCR was negative | ||
'''tphA3''' | '''tphA3''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR_on_a_DNA_template PCR] on tphA3 isolated from ''C. testosteroni'' |
** Annealing temperature: 59 °C | ** Annealing temperature: 59 °C | ||
** [[Primer]]: tphA3-Prefix_F and tphA3-Suffix_R | ** [[Primer]]: tphA3-Prefix_F and tphA3-Suffix_R | ||
| - | * Both PCR products were purified via [ | + | * Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 238: | Line 238: | ||
| tphA3-prefix/suffix || 30.5 | | tphA3-prefix/suffix || 30.5 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of mutated tphA3-prefix/suffix with EcoRI and PstI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of mutated tphA3-prefix/suffix (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_4_.2804.-08.06.12.29 pSB1C3 (cut with EcoRI and PstI)] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation |
** The PCR was positive | ** The PCR was positive | ||
* [[Inoculation]] of 10 mL of [[LB-medium]]-chloramphenicol with colony 1 and [[incubation]] | * [[Inoculation]] of 10 mL of [[LB-medium]]-chloramphenicol with colony 1 and [[incubation]] | ||
* [[Miniprep]] of the culture and a [[glycerine stock]] was made | * [[Miniprep]] of the culture and a [[glycerine stock]] was made | ||
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 252: | Line 252: | ||
| pSB1C3-tphA3-prefix/suffix || 79.6 | | pSB1C3-tphA3-prefix/suffix || 79.6 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of pSB1C3-tphA3-prefix/suffix with EcoRI and PstI |
* Preparation for [[sequencing]] | * Preparation for [[sequencing]] | ||
** Sequence was confirmed | ** Sequence was confirmed | ||
'''tphB''' | '''tphB''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR_on_a_DNA_template PCR] on tphB isolated from ''C. testosteroni'' |
** Annealing temperature: 59 °C | ** Annealing temperature: 59 °C | ||
** [[Primer]]: tphB-Prefix and tphB-Suffix_R | ** [[Primer]]: tphB-Prefix and tphB-Suffix_R | ||
| - | * Both PCR products were purified via [ | + | * Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 267: | Line 267: | ||
| tphB_prefix/suffix || 20.3 | | tphB_prefix/suffix || 20.3 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of mutated tphB-prefix/suffix with EcoRI and PstI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of mutated tphB-prefix/suffix (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_4_.2804.-08.06.12.29 pSB1C3 (cut with EcoRI and PstI)] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation |
** The PCR was negative | ** The PCR was negative | ||
| Line 276: | Line 276: | ||
'''tphB''' | '''tphB''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR_on_a_DNA_template PCR] on tphB isolated from ''C. testosteroni'' |
** Annealing temperature: 59 °C | ** Annealing temperature: 59 °C | ||
** [[Primer]]: tphB-Prefix and tphB-Suffix_R | ** [[Primer]]: tphB-Prefix and tphB-Suffix_R | ||
| - | * Both PCR products were purified via [ | + | * Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 287: | Line 287: | ||
| tphB_prefix/suffix || 52.5 | | tphB_prefix/suffix || 52.5 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of mutated tphB-prefix/suffix with EcoRI and PstI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of mutated tphB-prefix/suffix (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_4_.2804.-08.06.12.29 pSB1C3 (cut with EcoRI and PstI)] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation |
** The PCR was positive | ** The PCR was positive | ||
* [[Inoculation]] of 10 mL of [[LB-medium]]-chloramphenicol with colony 1 and [[incubation]] | * [[Inoculation]] of 10 mL of [[LB-medium]]-chloramphenicol with colony 1 and [[incubation]] | ||
* [[Miniprep]] of the culture and a [[glycerine stock]] was made | * [[Miniprep]] of the culture and a [[glycerine stock]] was made | ||
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 301: | Line 301: | ||
| pSB1C3-tphB-prefix/suffix || 35.8 | | pSB1C3-tphB-prefix/suffix || 35.8 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of pSB1C3-tphB-prefix/suffix with EcoRI and PstI |
* Preparation for [[sequencing]] | * Preparation for [[sequencing]] | ||
** Sequence was confirmed | ** Sequence was confirmed | ||
| Line 307: | Line 307: | ||
==week 12 (30.07.-03.08.12)== | ==week 12 (30.07.-03.08.12)== | ||
'''tphA1''' | '''tphA1''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR_on_a_DNA_template PCR] on mutated tphA1 |
** Annealing temperature: 59 °C | ** Annealing temperature: 59 °C | ||
** [[Primer]]: tphA1-Suffix_R and tphA1-l-Prefix | ** [[Primer]]: tphA1-Suffix_R and tphA1-l-Prefix | ||
| - | * Both PCR products were purified via [ | + | * Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 318: | Line 318: | ||
| Mutated tphA1-prefix/suffix || 34.2 | | Mutated tphA1-prefix/suffix || 34.2 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of mutated tphA1-prefix/suffix with EcoRI and PstI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of mutated tphA1-prefix/suffix (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_4_.2804.-08.06.12.29 pSB1C3 (cut with EcoRI and PstI)] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation |
** The PCR was positive | ** The PCR was positive | ||
| Line 328: | Line 328: | ||
* [[Inoculation]] of 10 mL of [[LB-medium]]-chloramphenicol with colony 1 and [[incubation]] | * [[Inoculation]] of 10 mL of [[LB-medium]]-chloramphenicol with colony 1 and [[incubation]] | ||
* [[Miniprep]] of the culture and a [[glycerine stock]] was made | * [[Miniprep]] of the culture and a [[glycerine stock]] was made | ||
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 335: | Line 335: | ||
| pSB1C3-tphA1 || 60.5 | | pSB1C3-tphA1 || 60.5 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of pSB1C3-tphA1-prefix/suffix with EcoRI and PstI |
[[File:WIKI-2012-08-02_pSB1C3-tphA1_test_restriction.jpg|thumb|none|x300px|alt=A|Test restriciton digest of psB1C3-tphA1 with EcoRI and PstI (GeneRuler 100bp Plus DNA Ladder, Fermentas)]] | [[File:WIKI-2012-08-02_pSB1C3-tphA1_test_restriction.jpg|thumb|none|x300px|alt=A|Test restriciton digest of psB1C3-tphA1 with EcoRI and PstI (GeneRuler 100bp Plus DNA Ladder, Fermentas)]] | ||
| Line 346: | Line 346: | ||
'''tphA2''' | '''tphA2''' | ||
* Reconstitution of the tphA2 gene synthesis | * Reconstitution of the tphA2 gene synthesis | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of the tphA2 gene synthesis |
* Inoculation of 10 mL [[LB-medium]]-kanamycin with one colony of the transformation and [[incubation]] | * Inoculation of 10 mL [[LB-medium]]-kanamycin with one colony of the transformation and [[incubation]] | ||
* [[Miniprep]] of the culture | * [[Miniprep]] of the culture | ||
| Line 356: | Line 356: | ||
|} | |} | ||
*[[Restriktion digest]] of the tphA2 gene synthesis with EcoRI and PstI | *[[Restriktion digest]] of the tphA2 gene synthesis with EcoRI and PstI | ||
| - | *[ | + | *[https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of the tphA2 gene synthesis and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_4_.2804.-08.06.12.29 pSB1C3 (cut with EcoRI and PstI)] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation |
** The PCR was positive | ** The PCR was positive | ||
* [[Inoculation]] of 10 mL of [[LB-medium]]-chloramphenicol with colony XX and [[incubation]] | * [[Inoculation]] of 10 mL of [[LB-medium]]-chloramphenicol with colony XX and [[incubation]] | ||
* [[Miniprep]] of the culture and a [[glycerine stock]] was made | * [[Miniprep]] of the culture and a [[glycerine stock]] was made | ||
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 376: | Line 376: | ||
'''aroY''' | '''aroY''' | ||
* Reconstitution of the aroY gene synthesis | * Reconstitution of the aroY gene synthesis | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of the aroY gene synthesis |
* Inoculation of 10 mL [[LB-medium]]-kanamycin with one colony of the transformation and [[incubation]] | * Inoculation of 10 mL [[LB-medium]]-kanamycin with one colony of the transformation and [[incubation]] | ||
* [[Miniprep]] of the culture | * [[Miniprep]] of the culture | ||
| Line 386: | Line 386: | ||
|} | |} | ||
*[[Restriktion digest]] of the aroY gene synthesis with EcoRI and PstI | *[[Restriktion digest]] of the aroY gene synthesis with EcoRI and PstI | ||
| - | *[ | + | *[https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of the aroY gene synthesis and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_4_.2804.-08.06.12.29 pSB1C3 (cut with EcoRI and PstI)] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation |
** The PCR was negative | ** The PCR was negative | ||
'''Other''' | '''Other''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] digest of BBa_J23100 by EcoRI and PstI |
| - | ** Purification of plasmid backbone J61002 via [ | + | ** Purification of plasmid backbone J61002 via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | ** Concentrations measured by [ | + | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
::{| class="wikitable" | ::{| class="wikitable" | ||
|- | |- | ||
| Line 405: | Line 405: | ||
'''Other''' | '''Other''' | ||
* Designing primers for over expression and operon construction | * Designing primers for over expression and operon construction | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of pPR-IBA2 |
* [[Inoculation]] of 10 mL of [[LB-medium]]-chloramphenicol with one colony and [[incubation]] | * [[Inoculation]] of 10 mL of [[LB-medium]]-chloramphenicol with one colony and [[incubation]] | ||
* [[Midiprep]] of the culture and a [[glycerine stock]] was made | * [[Midiprep]] of the culture and a [[glycerine stock]] was made | ||
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 415: | Line 415: | ||
| pPR-IBA2 || 127 | | pPR-IBA2 || 127 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] digest of pPR-IBA2 with EcoRI and PstI |
| - | ** Purification of plasmid backbone pPR-IBA2 via [ | + | ** Purification of plasmid backbone pPR-IBA2 via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | ** Concentrations measured by [ | + | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
::{| class="wikitable" | ::{| class="wikitable" | ||
|- | |- | ||
| Line 428: | Line 428: | ||
===Operon construction=== | ===Operon construction=== | ||
'''tphA1''' | '''tphA1''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR_on_a_DNA_template PCR] on pSB1C3-tphA1 |
** Annealing temperature: 59 °C | ** Annealing temperature: 59 °C | ||
** [[Primer]]: RBS-tphA1 and Suffix | ** [[Primer]]: RBS-tphA1 and Suffix | ||
| - | * Both PCR products were purified via [ | + | * Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 439: | Line 439: | ||
| tphA1 with RBS || 33.5 | | tphA1 with RBS || 33.5 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of RBS-tphA1 with EcoRI and PstI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of RBS-tphA1 (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_14_.2813.-17.08.12.29 J61002 (cut with EcoRI and PstI)] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation |
** The PCR was positive | ** The PCR was positive | ||
* [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | * [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | ||
* [[Miniprep]] of the culture and a [[glycerine stock]] was made | * [[Miniprep]] of the culture and a [[glycerine stock]] was made | ||
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 454: | Line 454: | ||
|} | |} | ||
'''tphA2''' | '''tphA2''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR_on_a_DNA_template PCR] on pSB1C3-tphA2 |
** Annealing temperature: 59 °C | ** Annealing temperature: 59 °C | ||
** [[Primer]]: RBS-tphA2 and Suffix | ** [[Primer]]: RBS-tphA2 and Suffix | ||
| - | * Both PCR products were purified via [ | + | * Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 465: | Line 465: | ||
| tphA2 with RBS || 46.8 | | tphA2 with RBS || 46.8 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of RBS-tphA2 with EcoRI and PstI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of RBS-tphA2 (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_14_.2813.-17.08.12.29 J61002 (cut with EcoRI and PstI)] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation |
** The PCR was positive | ** The PCR was positive | ||
* [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | * [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | ||
* [[Miniprep]] of the culture and a [[glycerine stock]] was made | * [[Miniprep]] of the culture and a [[glycerine stock]] was made | ||
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 480: | Line 480: | ||
|} | |} | ||
'''tphA3''' | '''tphA3''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR_on_a_DNA_template PCR] on pSB1C3-tphA3 |
** Annealing temperature: 59 °C | ** Annealing temperature: 59 °C | ||
** [[Primer]]: RBS-tphA3 and Suffix | ** [[Primer]]: RBS-tphA3 and Suffix | ||
| - | * Both PCR products were purified via [ | + | * Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 491: | Line 491: | ||
| tphA3 with RBS || 26.5 | | tphA3 with RBS || 26.5 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of RBS-tphA3 with EcoRI and PstI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of RBS-tphA3 (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_14_.2813.-17.08.12.29 J61002 (cut with EcoRI and PstI)] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation |
** The PCR was positive | ** The PCR was positive | ||
* [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | * [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | ||
* [[Miniprep]] of the culture and a [[glycerine stock]] was made | * [[Miniprep]] of the culture and a [[glycerine stock]] was made | ||
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 506: | Line 506: | ||
|} | |} | ||
'''tphB''' | '''tphB''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR_on_a_DNA_template PCR] on pSB1C3-tphB |
** Annealing temperature: 59 °C | ** Annealing temperature: 59 °C | ||
** [[Primer]]: RBS-tphB and Suffix | ** [[Primer]]: RBS-tphB and Suffix | ||
| - | * Both PCR products were purified via [ | + | * Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 517: | Line 517: | ||
| tphB with RBS || 49.2 | | tphB with RBS || 49.2 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of RBS-tphB with EcoRI and PstI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of RBS-tphB (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_14_.2813.-17.08.12.29 J61002 (cut with EcoRI and PstI)] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation |
** The PCR was positive | ** The PCR was positive | ||
* [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | * [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | ||
* [[Miniprep]] of the culture and a [[glycerine stock]] was made | * [[Miniprep]] of the culture and a [[glycerine stock]] was made | ||
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 532: | Line 532: | ||
|} | |} | ||
'''aroY''' | '''aroY''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR_on_a_DNA_template PCR] on pSB1C3-aroY |
** Annealing temperature: 59 °C | ** Annealing temperature: 59 °C | ||
** [[Primer]]: RBS-aroY and Suffix | ** [[Primer]]: RBS-aroY and Suffix | ||
| - | * Both PCR products were purified via [ | + | * Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 543: | Line 543: | ||
| aroY with RBS || 55.2 | | aroY with RBS || 55.2 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of RBS-aroY with EcoRI and PstI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of RBS-aroY (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_14_.2813.-17.08.12.29 J61002 (cut with EcoRI and PstI)] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation |
** The PCR was positive | ** The PCR was positive | ||
* [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | * [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | ||
* [[Miniprep]] of the culture and a [[glycerine stock]] was made | * [[Miniprep]] of the culture and a [[glycerine stock]] was made | ||
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 561: | Line 561: | ||
===Over expression=== | ===Over expression=== | ||
'''tphA1''' | '''tphA1''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR_on_a_DNA_template PCR] on pSB1C3-tphA1 |
** Annealing temperature: 59 °C | ** Annealing temperature: 59 °C | ||
** [[Primer]]: EcoRIGFxa-tphA1 and Suffix | ** [[Primer]]: EcoRIGFxa-tphA1 and Suffix | ||
| - | * Both PCR products were purified via [ | + | * Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 572: | Line 572: | ||
| tphA1_over-ex || 116.2 | | tphA1_over-ex || 116.2 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of tphA1_over-ex with EcoRI and PstI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of tphA1_over-ex (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_15_.2820.-24.08.12.29 pPR-IBA2 (cut with EcoRI and PstI)] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation |
** The PCR was positive | ** The PCR was positive | ||
* [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | * [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | ||
* [[Miniprep]] of the culture and a [[glycerine stock]] was made | * [[Miniprep]] of the culture and a [[glycerine stock]] was made | ||
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 587: | Line 587: | ||
|} | |} | ||
'''tphA2''' | '''tphA2''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR_on_a_DNA_template PCR] on pSB1C3-tphA2 |
** Annealing temperature: 59 °C | ** Annealing temperature: 59 °C | ||
** [[Primer]]: EcoRIGFxa-tphA2 and Suffix | ** [[Primer]]: EcoRIGFxa-tphA2 and Suffix | ||
| - | * Both PCR products were purified via [ | + | * Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 598: | Line 598: | ||
| tphA2_over-ex || 63.9 | | tphA2_over-ex || 63.9 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of tphA2_over-ex with EcoRI and PstI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of tphA2_over-ex (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_15_.2820.-24.08.12.29 pPR-IBA2 (cut with EcoRI and PstI)] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation |
** The PCR was positive | ** The PCR was positive | ||
* [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | * [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | ||
* [[Miniprep]] of the culture and a [[glycerine stock]] was made | * [[Miniprep]] of the culture and a [[glycerine stock]] was made | ||
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 613: | Line 613: | ||
|} | |} | ||
'''tphA3''' | '''tphA3''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR_on_a_DNA_template PCR] on pSB1C3-tphA3 |
** Annealing temperature: 59 °C | ** Annealing temperature: 59 °C | ||
** [[Primer]]: EcoRIGFxa-tphA3 and Suffix | ** [[Primer]]: EcoRIGFxa-tphA3 and Suffix | ||
| - | * Both PCR products were purified via [ | + | * Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 624: | Line 624: | ||
| tphA3_over-ex || 90.4 | | tphA3_over-ex || 90.4 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of tphA3_over-ex with EcoRI and PstI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of tphA3_over-ex (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_15_.2820.-24.08.12.29 pPR-IBA2 (cut with EcoRI and PstI)] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation |
** The PCR was positive | ** The PCR was positive | ||
* [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | * [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | ||
* [[Miniprep]] of the culture and a [[glycerine stock]] was made | * [[Miniprep]] of the culture and a [[glycerine stock]] was made | ||
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 639: | Line 639: | ||
|} | |} | ||
'''tphB''' | '''tphB''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR_on_a_DNA_template PCR] on pSB1C3-tphB |
** Annealing temperature: 59 °C | ** Annealing temperature: 59 °C | ||
** [[Primer]]: EcoRIGFxa-tphB and Suffix | ** [[Primer]]: EcoRIGFxa-tphB and Suffix | ||
| - | * Both PCR products were purified via [ | + | * Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 650: | Line 650: | ||
| tphB_over-ex || 87.5 | | tphB_over-ex || 87.5 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of tphB_over-ex with EcoRI and PstI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of tphB_over-ex (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_15_.2820.-24.08.12.29 pPR-IBA2 (cut with EcoRI and PstI)] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation |
** The PCR was positive | ** The PCR was positive | ||
* [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | * [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | ||
* [[Miniprep]] of the culture and a [[glycerine stock]] was made | * [[Miniprep]] of the culture and a [[glycerine stock]] was made | ||
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 665: | Line 665: | ||
|} | |} | ||
'''aroY''' | '''aroY''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR_on_a_DNA_template PCR] on pSB1C3-aroY |
** Annealing temperature: 59 °C | ** Annealing temperature: 59 °C | ||
** [[Primer]]: EcoRIGFxa-aroY and Suffix | ** [[Primer]]: EcoRIGFxa-aroY and Suffix | ||
| - | * Both PCR products were purified via [ | + | * Both PCR products were purified via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 676: | Line 676: | ||
| aroY_over-ex || 105.1 | | aroY_over-ex || 105.1 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of aroY_over-ex with EcoRI and PstI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of aroY_over-ex (cut with EcoRI and PstI) and [https://2012.igem.org/Team:TU_Darmstadt/Labjournal/Metabolism#week_15_.2820.-24.08.12.29 pPR-IBA2 (cut with EcoRI and PstI)] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] for verification of the transformation |
** The PCR was positive | ** The PCR was positive | ||
* [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | * [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | ||
* [[Miniprep]] of the culture and a [[glycerine stock]] was made | * [[Miniprep]] of the culture and a [[glycerine stock]] was made | ||
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 694: | Line 694: | ||
===Operon construction=== | ===Operon construction=== | ||
'''RBS-tphA1-RBS-tphA2''' | '''RBS-tphA1-RBS-tphA2''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] digest of J61002-RBS-tphA1 by EcoRI and SpeI |
| - | * Purification of insert RBS-tphA1 RBS-tphA1 (cut with EcoRI and SpeI) via [ | + | * Purification of insert RBS-tphA1 RBS-tphA1 (cut with EcoRI and SpeI) via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | ** Concentrations measured by [ | + | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
::{| class="wikitable" | ::{| class="wikitable" | ||
|- | |- | ||
| Line 703: | Line 703: | ||
| RBS-tphA1 (cut with EcoRI and SpeI)|| 50.2 | | RBS-tphA1 (cut with EcoRI and SpeI)|| 50.2 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of J61002-RBS-tphA2 EcoRI and XbaI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Dephosphorylation Dephosphorylation] of the plasmid backbone J61002-RBS-tphA2 (cut with EcoRI and XbaI) |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of the plasmid backbone J61002-RBS-tphA2 (cut with EcoRI and XbaI)and RBS-tphA1 (cut with EcoRI and SpeI) |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of the ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] of the transformation for verification |
** The PCR was positive | ** The PCR was positive | ||
| Line 714: | Line 714: | ||
* [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony 4 and [[incubation]] | * [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony 4 and [[incubation]] | ||
* [[Miniprep]] of the culture and a [[glycerine stock]] was made | * [[Miniprep]] of the culture and a [[glycerine stock]] was made | ||
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 722: | Line 722: | ||
|} | |} | ||
'''RBS-tphA3-RBS-tphB''' | '''RBS-tphA3-RBS-tphB''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] digest of J61002-RBS-tphA3 by EcoRI and SpeI |
| - | * Purification of insert RBS-tphA3 (cut with EcoRI and SpeI) via [ | + | * Purification of insert RBS-tphA3 (cut with EcoRI and SpeI) via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | ** Concentrations measured by [ | + | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
::{| class="wikitable" | ::{| class="wikitable" | ||
|- | |- | ||
| Line 731: | Line 731: | ||
| RBS-tphA3 (cut with EcoRI and SpeI)|| 178.9 | | RBS-tphA3 (cut with EcoRI and SpeI)|| 178.9 | ||
|} | |} | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of J61002-RBS-tphB EcoRI and XbaI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Dephosphorylation Dephosphorylation] of the plasmid backbone J61002-RBS-tphB (cut with EcoRI and XbaI) |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of the plasmid backbone J61002-RBS-tphB (cut with EcoRI and XbaI)and RBS-tphA3 (cut with EcoRI and SpeI) |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of the ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] of the transformation for verification |
** The PCR was positive | ** The PCR was positive | ||
* [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | * [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony XXX and [[incubation]] | ||
* [[Miniprep]] of the culture and a [[glycerine stock]] was made | * [[Miniprep]] of the culture and a [[glycerine stock]] was made | ||
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 749: | Line 749: | ||
'''RBS-tphA1-RBS-tphA2-RBS-tphA3-RBS-tphB''' | '''RBS-tphA1-RBS-tphA2-RBS-tphA3-RBS-tphB''' | ||
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of J61002-RBS-tphA1-RBS-tphA2 by EcoRI and SpeI |
| - | * Purification of insert RBS-tphA1-RBS-tphA2 (cut with EcoRI and SpeI) via [ | + | * Purification of insert RBS-tphA1-RBS-tphA2 (cut with EcoRI and SpeI) via [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Gel_and_PCR_Clean-Up gel extraction] |
| - | ** Concentrations measured by [ | + | ** Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
::{| class="wikitable" | ::{| class="wikitable" | ||
|- | |- | ||
| Line 759: | Line 759: | ||
|} | |} | ||
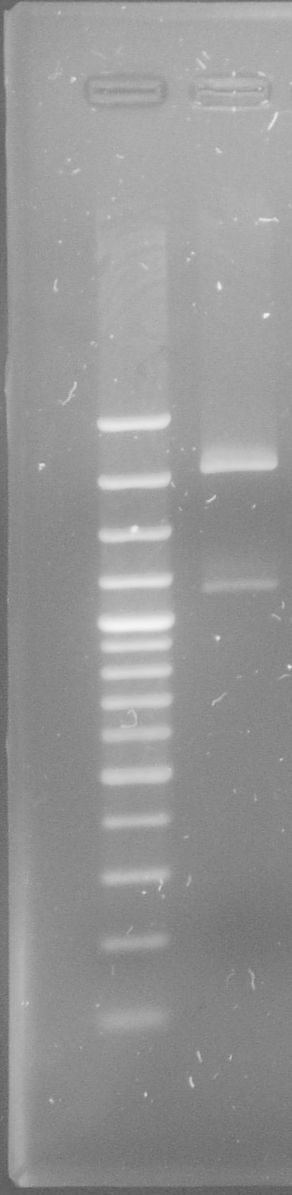
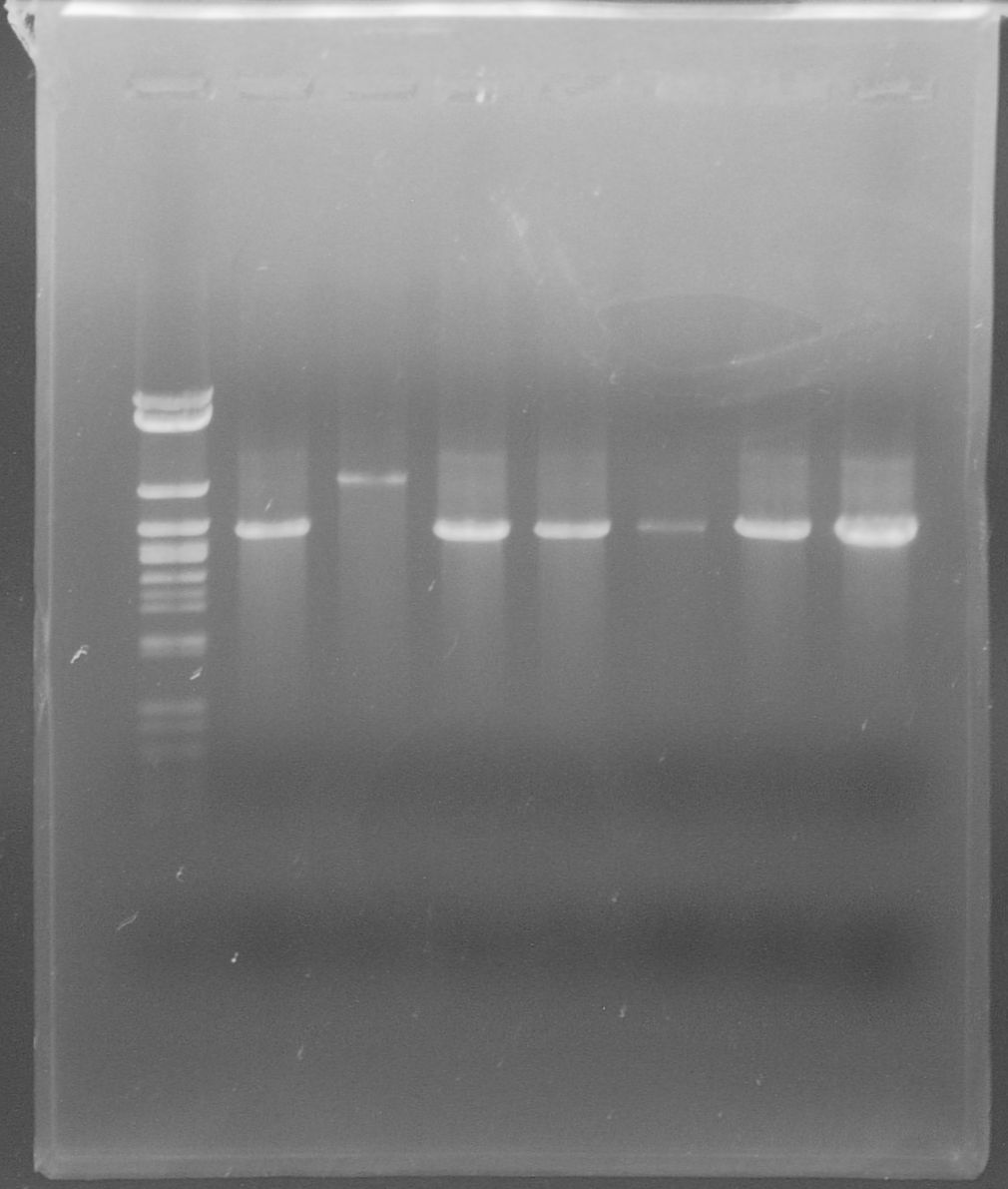
| - | [[File:WIKI-2012-09-04_E%2BS_restriction_RBS_tphA1tphA2.jpg|thumb|none|alt=A| | + | [[File:WIKI-2012-09-04_E%2BS_restriction_RBS_tphA1tphA2.jpg|thumb|none|alt=A|restriction of of J61002-RBS-tphA1-RBS-tphA2 by EcoRI and SpeI (Lambda DNA/Eco47I (AvaII) Marker, 13, Fermentas)]] |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Restriction_digest restriction] of J61002-RBS-tphA3-RBS-tphB EcoRI and XbaI |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Dephosphorylation Dephosphorylation] of the plasmid backbone J61002-RBS-tphA3-RBS-tphB (cut with EcoRI and XbaI) |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Ligation Ligation] of the plasmid backbone J61002-RBS-tphA3-RBS-tphB EcoRI (cut with EcoRI and XbaI)and RBS-tphA1-RBS-tphA2 (cut with EcoRI and SpeI) |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Heat_shock_transformation_with_E._coli Transformation] of the ligation mix |
| - | * [ | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#PCR colony-PCR] of the transformation for verification |
** The PCR was positive | ** The PCR was positive | ||
| Line 772: | Line 772: | ||
* [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony 2 and [[incubation]] | * [[Inoculation]] of 10 mL of [[LB-medium]]-ampicillin with colony 2 and [[incubation]] | ||
* [[Miniprep]] of the culture and a [[glycerine stock]] was made | * [[Miniprep]] of the culture and a [[glycerine stock]] was made | ||
| - | * Concentrations measured by [ | + | * Concentrations measured by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Nanodrop Nanodrop] |
:{| class="wikitable" | :{| class="wikitable" | ||
|- | |- | ||
| Line 784: | Line 784: | ||
'''tphA2''' | '''tphA2''' | ||
* [[Inoculate]] 10 mL of [[LB-medium]]-ampicillin with pPR-IBA2-tphA1_over-ex and [[incubate]] | * [[Inoculate]] 10 mL of [[LB-medium]]-ampicillin with pPR-IBA2-tphA1_over-ex and [[incubate]] | ||
| - | * Over expression according to standard [ | + | * Over expression according to standard [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_overexpression protocol] |
'''tphB''' | '''tphB''' | ||
* [[Inoculate]] 10 mL of [[LB-medium]]-ampicillin with pPR-IBA2-tphA1_over-ex and [[incubate]] | * [[Inoculate]] 10 mL of [[LB-medium]]-ampicillin with pPR-IBA2-tphA1_over-ex and [[incubate]] | ||
| - | * Over expression according to standard [ | + | * Over expression according to standard [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_overexpression protocol] |
'''aroY''' | '''aroY''' | ||
* [[Inoculate]] 10 mL of [[LB-medium]]-ampicillin with pPR-IBA2-tphA1_over-ex and [[incubate]] | * [[Inoculate]] 10 mL of [[LB-medium]]-ampicillin with pPR-IBA2-tphA1_over-ex and [[incubate]] | ||
| - | * Over expression according to standard [ | + | * Over expression according to standard [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_overexpression protocol] |
'''tphA1''' | '''tphA1''' | ||
* [[Inoculate]] 10 mL of [[LB-medium]]-ampicillin with pPR-IBA2-tphA1_over-ex and [[incubate]] | * [[Inoculate]] 10 mL of [[LB-medium]]-ampicillin with pPR-IBA2-tphA1_over-ex and [[incubate]] | ||
| - | * Over expression according to standard [ | + | * Over expression according to standard [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_overexpression protocol] |
'''tphA3''' | '''tphA3''' | ||
* [[Inoculate]] 10 mL of [[LB-medium]]-ampicillin with pPR-IBA2-tphA1_over-ex and [[incubate]] | * [[Inoculate]] 10 mL of [[LB-medium]]-ampicillin with pPR-IBA2-tphA1_over-ex and [[incubate]] | ||
| - | * Over expression according to standard [ | + | * Over expression according to standard [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_overexpression protocol] |
'''SDS-PAGE of tphB overexpression and tphA2 overexpression respectively''' | '''SDS-PAGE of tphB overexpression and tphA2 overexpression respectively''' | ||
| - | * SDS-Page according to standard [ | + | * SDS-Page according to standard [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#SDS-PAGE protocol] |
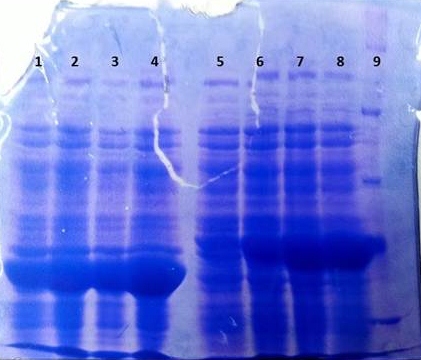
[[File:TphB_tphA2_overex.jpg|thumb|none|alt=A|Results of the overexpression tphA2/tphB]] | [[File:TphB_tphA2_overex.jpg|thumb|none|alt=A|Results of the overexpression tphA2/tphB]] | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 829: | Line 829: | ||
'''SDS-PAGE of overexpression from all five genes''' | '''SDS-PAGE of overexpression from all five genes''' | ||
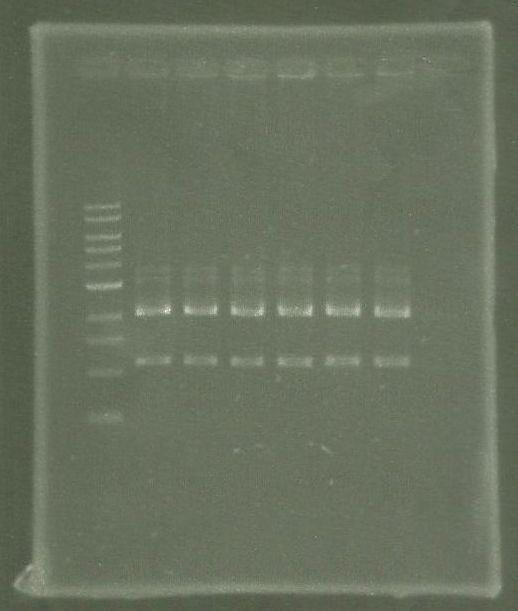
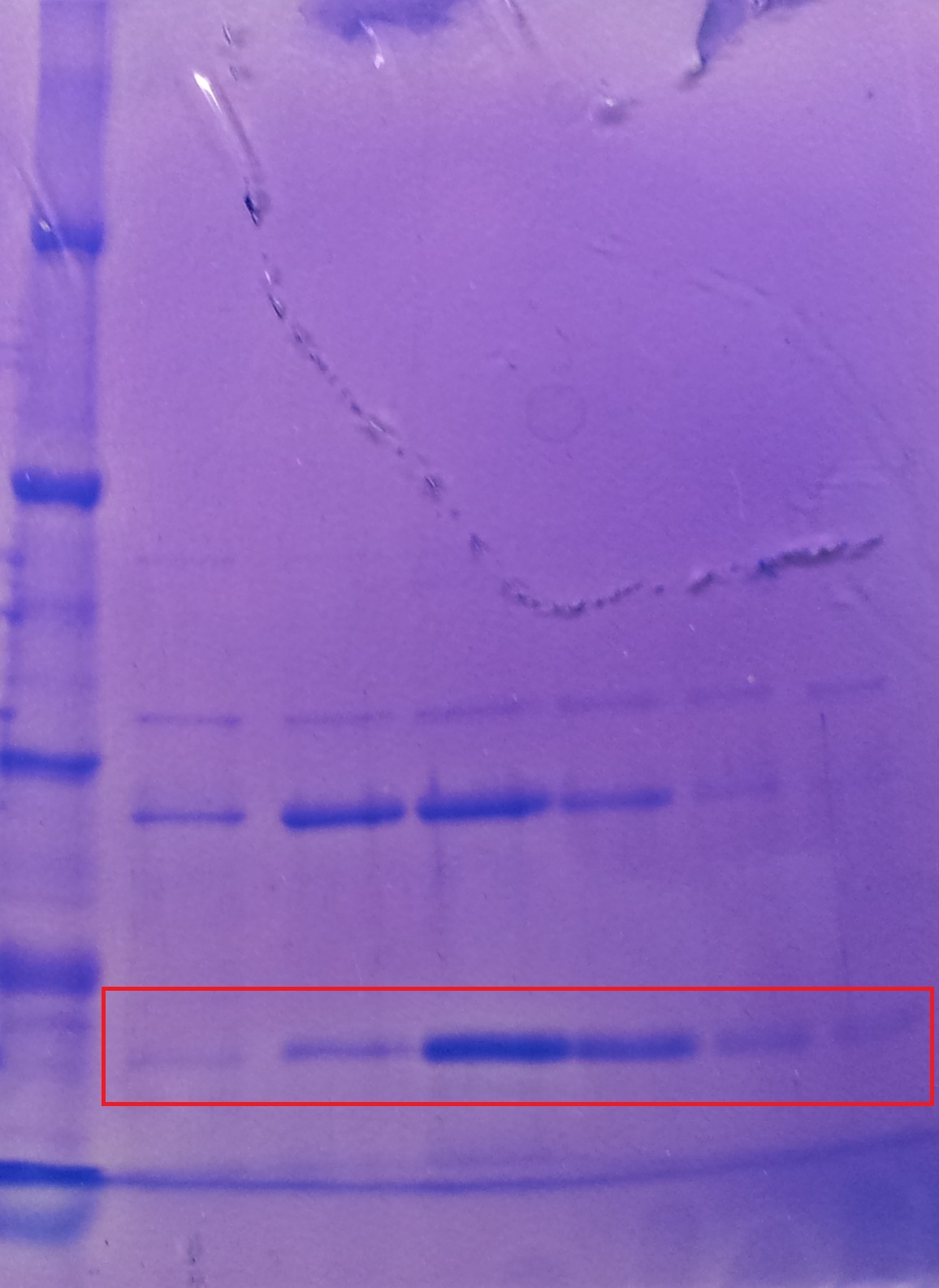
| - | * SDS-Page according to standard [ | + | * SDS-Page according to standard [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#SDS-PAGE protocol] |
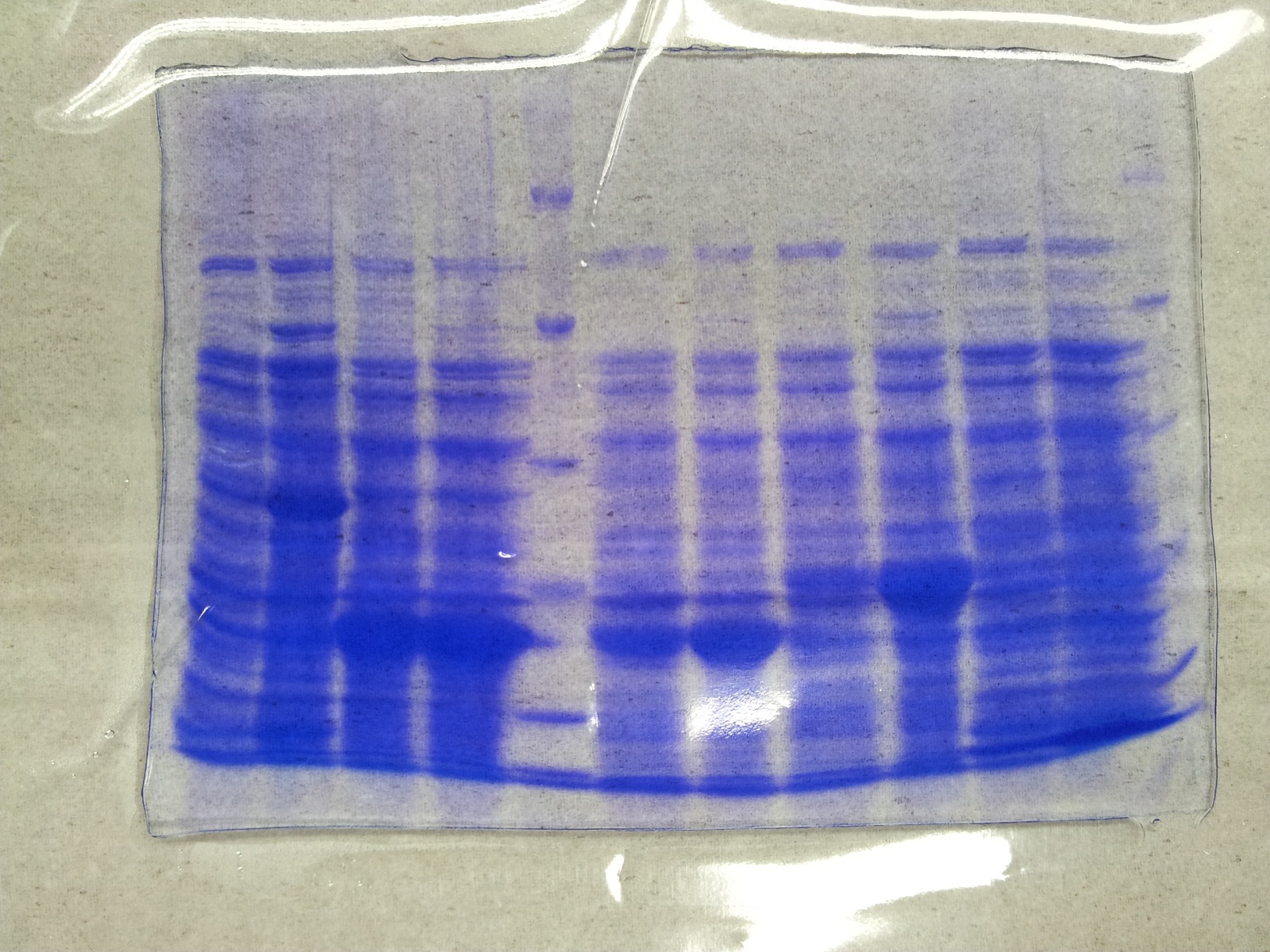
[[File: Overex_all.jpg |thumb|none|alt=A|Results of the overexpression aroY/tphA3/tphA1/tphA2/tphB/]] | [[File: Overex_all.jpg |thumb|none|alt=A|Results of the overexpression aroY/tphA3/tphA1/tphA2/tphB/]] | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 862: | Line 862: | ||
==week 18 (10.-17.09.12)== | ==week 18 (10.-17.09.12)== | ||
'''Purification of aroY''' | '''Purification of aroY''' | ||
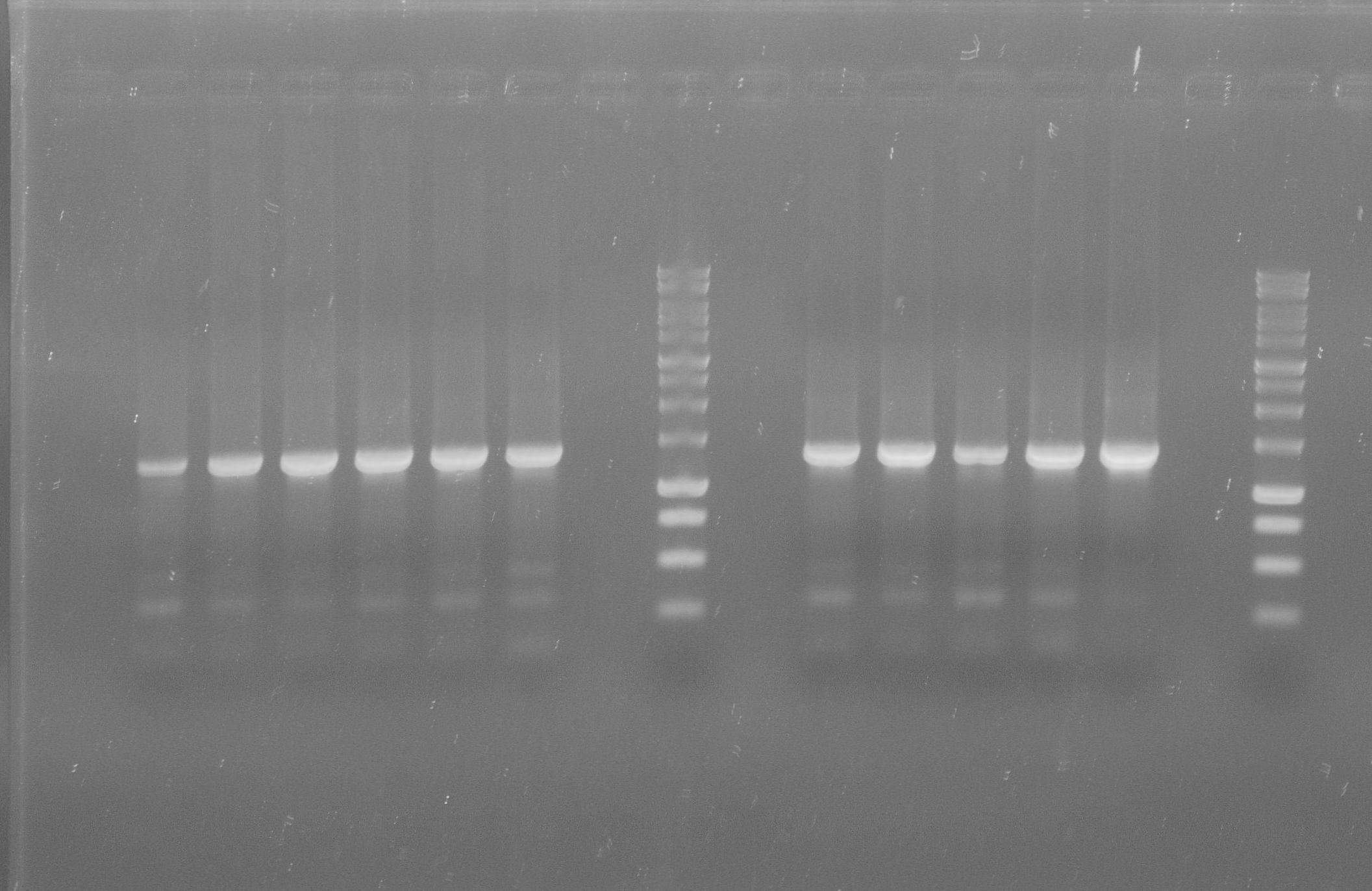
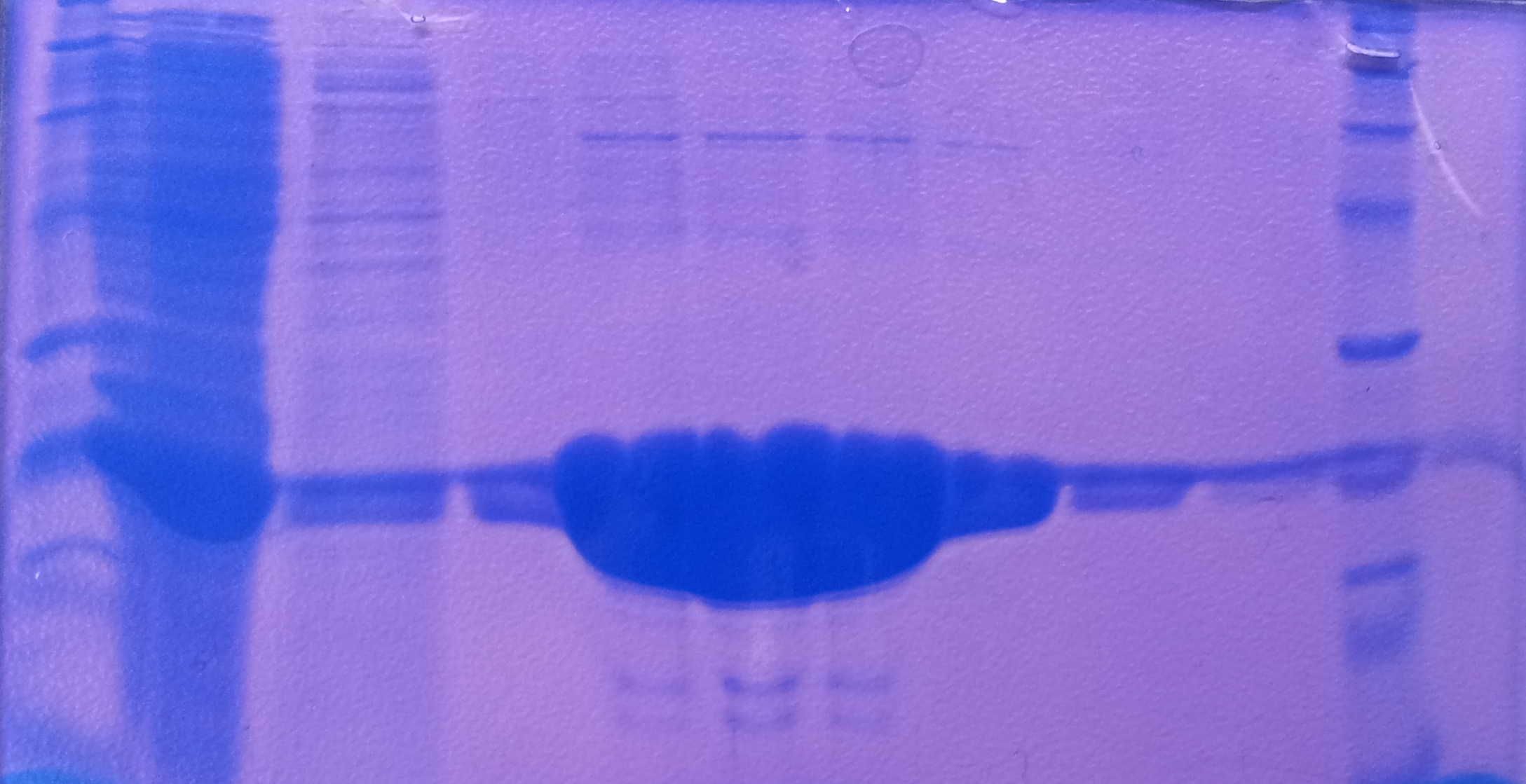
| - | * Protein purification according to | + | * Protein purification according to standard strep-tag [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_purification purification protocol ] |
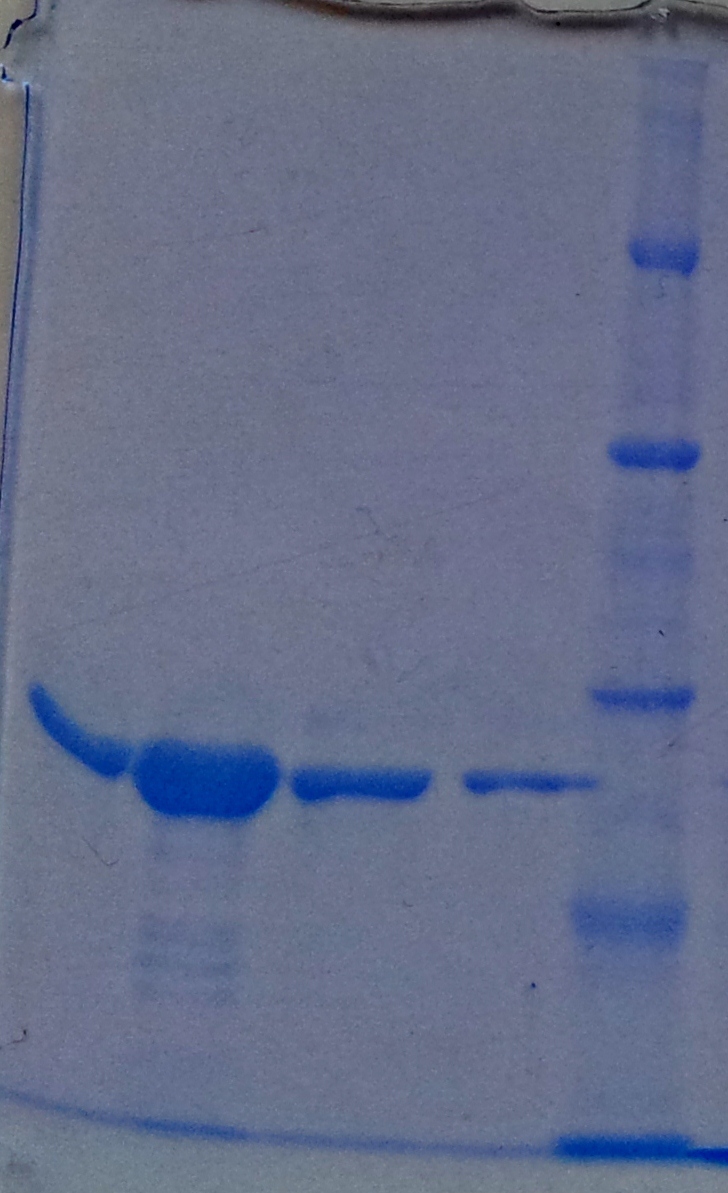
[[File:Aro_purificate.jpg|thumb|none|alt=A|Fractions of the aroY purfication]] | [[File:Aro_purificate.jpg|thumb|none|alt=A|Fractions of the aroY purfication]] | ||
| Line 882: | Line 882: | ||
'''Purification of TphA3''' | '''Purification of TphA3''' | ||
| - | * Protein purification according to | + | * Protein purification according to standard strep-tag [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_purification purification protocol ] |
[[File:Puri_A3.jpg|thumb|none|alt=A|Fractions of the TphA3 purfication]] | [[File:Puri_A3.jpg|thumb|none|alt=A|Fractions of the TphA3 purfication]] | ||
| Line 914: | Line 914: | ||
'''Purification of TphA1''' | '''Purification of TphA1''' | ||
| - | * Protein purification according to | + | * Protein purification according to standard strep-tag [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Metabolism#Protein_purification purification protocol ] |
[[File:Puri_A1.jpg|thumb|none|alt=A|Fractions of the TphA1 purfication]] | [[File:Puri_A1.jpg|thumb|none|alt=A|Fractions of the TphA1 purfication]] | ||
Revision as of 08:49, 23 September 2012
Protocols Metabolism
week 1 (14.-18.05.12)
Other
- Reconstitution of C. testosteroni KF-1 according to DSMZ protocol
- Cultivation of C. testosteroni KF-1 on agar plates with Medium 1
- Production of chemically competent E. coli DH5α and E. coli BL21(DE3)pLysS cells
week 2 (21.-25.05.12)
tphA1
- Isolation of the gene from C. testosteroni KF-1 genome using colony-PCR
- Annealing temperature: 49 °C
- Primer: tphA1-l-F and tphA1-l-R
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphA1 0.6
tphA3
- Isolation of the gene from C. testosteroni KF-1 genome using colony-PCR
- Annealing temperature: 59 °C
- Primer: tphA3-l-F and tphA3-l-R
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphA3 0.1
tphB
- Isolation of the gene from C. testosteroni KF-1 genome using colony-PCR
- Annealing temperature: 59 °C
- Primer: tphB-l-F and tphB-l-R
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphB 0.1
Other
- Transformation and midi prep of all used Biobricks
- Concentrations measured by Nanodrop
Biobrick Concentration [ng/µl] BBa_K316003 114.9 BBa_J23100 450.2 BBa_B0015 314.1 BBa_J61101 86.1
week 3 (28.05.-01.06.12)
tphA1
- Two PCRs were performed to mutate the PstI site in the wild type gene. The primer tphA1-l-PstI(99)-R and tphA1-l-PstI(99)-F respectively introduced a BsaI site
- tphA1 fragment 1
- PCR on tphA1 isolated from C. testosteroni
- Annealing temperature: 69 °C
- Primer: tphA1-l-PstI(99)-R and tphA1-l-R
- tphA1 fragment 2
- PCR on tphA1 isolated from C. testosteroni
- Annealing temperature: 69 °C
- Primer: tphA1-l-PstI(99)-F and tphA1-l-F
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] Fragment 1 40.3 Fragment 2 62.1
- Both fragments were cut with BsaI in a restriction
- The ligation mix differed from our standard protocol in the following manner
- 100 ng of fragment 1
- 200 ng of fragment 2
- 2 µL of 10x reaction buffer
- 1 µL of T4 DNA ligase
- add DI water up to 20 µL
- incubate for 15 minutes at 37 °C
- PCR on ligation mix
- Annealing temperature: 59 °C
- Primer: tphA1-l-R and tphA1-l-F
- The PCR product was purified via gel extraction
- Concentrations measured by Nanodrop
- The ligation mix differed from our standard protocol in the following manner
PCR product Concentration [ng/µl] Mutated tphA1 86.1
week 4 (04.-08.06.12)
Other
- restriction digest of BBa_K316003 by EcoRI and PstI
- Purification of plasmid backbone pSB1C3 via gel extraction
- Concentrations measured by Nanodrop
Plamid backbone Concentration [ng/µl] pSB1C3 42.6
- restriction digest of BBa_K316003 by XbaI and PstI
- Purification of insert xylE-dT via gel extraction
- Concentrations measured by Nanodrop
Insert Concentration [ng/µl] xylE-dT 22.2
week 5 (11.-15-06.12)
Other
- restriction digest of BBa_J23100 by SpeI and PstI
- Dephosphorylation of the restriction
- Ligation of BBa_J23100 (cut with SpeI and PstI) and xylE-dT (cut with XbaI and PstI)
- Transformation of the ligation mix
- colony-PCR of the transformation for verification
- After overnight incubation of colony x in LB medium with ampicilin a glycerine stock was made
week 6 (18.-22.06.12)
- No work progress
week 7 (25.-29.06.12)
Other
- Funktional testing of BBa_J23100-xylE-dT
- We inoculated 50 mL LB-medium-ampicilin with 10 µl of the glycerine stock BBa_J23100-xylE-dT
- After incubation we centrifuged the culture at 4600x g for 10 minutes
- We resuspended the pellet with the 1000 µL pipette in 3 mL PBS buffer and added PBS to 120 ml
- We added 2 mL of 0.5 M catechol solution to the cell suspension
- We observed a colour change colourless to light yellow
week 8 (02.-06.07.12)
- No work progress
week 9 (09.-13.07.12)
Other
- Designing primers with prefix and suffix respectively
- Designing genes (aroY and tphA2 respectivley) according to the biobrick standard for gene synthesis. Gene synthesis was performed by GeneArt®
week 10 (16.-20.07.12)
tphA1
- PCR on mutated tphA1
- Annealing temperature: 59 °C
- Primer: tphA1-Suffix_R and tphA1-l-Prefix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] Mutated tphA1-prefix/suffix 62.0
- restriction of mutated tphA1-prefix/suffix with EcoRI and PstI
- Ligation of mutated tphA1-prefix/suffix (cut with EcoRI and PstI) and pSB1C3 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was negative
tphA3
- PCR on tphA3 isolated from C. testosteroni
- Annealing temperature: 59 °C
- Primer: tphA3-Prefix_F and tphA3-Suffix_R
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphA3-prefix/suffix 30.5
- restriction of mutated tphA3-prefix/suffix with EcoRI and PstI
- Ligation of mutated tphA3-prefix/suffix (cut with EcoRI and PstI) and pSB1C3 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-medium-chloramphenicol with colony 1 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] pSB1C3-tphA3-prefix/suffix 79.6
- restriction of pSB1C3-tphA3-prefix/suffix with EcoRI and PstI
- Preparation for sequencing
- Sequence was confirmed
tphB
- PCR on tphB isolated from C. testosteroni
- Annealing temperature: 59 °C
- Primer: tphB-Prefix and tphB-Suffix_R
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphB_prefix/suffix 20.3
- restriction of mutated tphB-prefix/suffix with EcoRI and PstI
- Ligation of mutated tphB-prefix/suffix (cut with EcoRI and PstI) and pSB1C3 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was negative
week 11 (23.-27.07.12)
tphB
- PCR on tphB isolated from C. testosteroni
- Annealing temperature: 59 °C
- Primer: tphB-Prefix and tphB-Suffix_R
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphB_prefix/suffix 52.5
- restriction of mutated tphB-prefix/suffix with EcoRI and PstI
- Ligation of mutated tphB-prefix/suffix (cut with EcoRI and PstI) and pSB1C3 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-medium-chloramphenicol with colony 1 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] pSB1C3-tphB-prefix/suffix 35.8
- restriction of pSB1C3-tphB-prefix/suffix with EcoRI and PstI
- Preparation for sequencing
- Sequence was confirmed
week 12 (30.07.-03.08.12)
tphA1
- PCR on mutated tphA1
- Annealing temperature: 59 °C
- Primer: tphA1-Suffix_R and tphA1-l-Prefix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] Mutated tphA1-prefix/suffix 34.2
- restriction of mutated tphA1-prefix/suffix with EcoRI and PstI
- Ligation of mutated tphA1-prefix/suffix (cut with EcoRI and PstI) and pSB1C3 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-medium-chloramphenicol with colony 1 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] pSB1C3-tphA1 60.5
- restriction of pSB1C3-tphA1-prefix/suffix with EcoRI and PstI
- Preparation for sequencing
- Sequence was confirmed
week 13 (06.-10.08.12)
tphA2
- Reconstitution of the tphA2 gene synthesis
- Transformation of the tphA2 gene synthesis
- Inoculation of 10 mL LB-medium-kanamycin with one colony of the transformation and incubation
- Miniprep of the culture
Miniprep Concentration [ng/µl] tphA2 gene synthesis 112.6
- Restriktion digest of the tphA2 gene synthesis with EcoRI and PstI
- Ligation of the tphA2 gene synthesis and pSB1C3 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-medium-chloramphenicol with colony XX and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] pSB1C3-tphA2-prefix/suffix 111.1
- Preparation for sequencing
- Sequence was confirmed
week 14 (13.-17.08.12)
aroY
- Reconstitution of the aroY gene synthesis
- Transformation of the aroY gene synthesis
- Inoculation of 10 mL LB-medium-kanamycin with one colony of the transformation and incubation
- Miniprep of the culture
Miniprep Concentration [ng/µl] aroY gene synthesis 63.25
- Restriktion digest of the aroY gene synthesis with EcoRI and PstI
- Ligation of the aroY gene synthesis and pSB1C3 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was negative
Other
- restriction digest of BBa_J23100 by EcoRI and PstI
- Purification of plasmid backbone J61002 via gel extraction
- Concentrations measured by Nanodrop
Plamid backbone Concentration [ng/µl] J61002 42.5
week 15 (20.-24.08.12)
Other
- Designing primers for over expression and operon construction
- Transformation of pPR-IBA2
- Inoculation of 10 mL of LB-medium-chloramphenicol with one colony and incubation
- Midiprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Midiprep Concentration [ng/µl] pPR-IBA2 127
- restriction digest of pPR-IBA2 with EcoRI and PstI
- Purification of plasmid backbone pPR-IBA2 via gel extraction
- Concentrations measured by Nanodrop
Plamid backbone Concentration [ng/µl] pPR-IBA2 35.6
week 16 (27.-31.08.12)
Operon construction
tphA1
- PCR on pSB1C3-tphA1
- Annealing temperature: 59 °C
- Primer: RBS-tphA1 and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphA1 with RBS 33.5
- restriction of RBS-tphA1 with EcoRI and PstI
- Ligation of RBS-tphA1 (cut with EcoRI and PstI) and J61002 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-medium-ampicillin with colony XXX and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] J61002-RBS-tphA1 79,6
tphA2
- PCR on pSB1C3-tphA2
- Annealing temperature: 59 °C
- Primer: RBS-tphA2 and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphA2 with RBS 46.8
- restriction of RBS-tphA2 with EcoRI and PstI
- Ligation of RBS-tphA2 (cut with EcoRI and PstI) and J61002 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-medium-ampicillin with colony XXX and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] J61002-RBS-tphA2 80.3
tphA3
- PCR on pSB1C3-tphA3
- Annealing temperature: 59 °C
- Primer: RBS-tphA3 and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphA3 with RBS 26.5
- restriction of RBS-tphA3 with EcoRI and PstI
- Ligation of RBS-tphA3 (cut with EcoRI and PstI) and J61002 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-medium-ampicillin with colony XXX and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] J61002-RBS-tphA3 67.5
tphB
- PCR on pSB1C3-tphB
- Annealing temperature: 59 °C
- Primer: RBS-tphB and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphB with RBS 49.2
- restriction of RBS-tphB with EcoRI and PstI
- Ligation of RBS-tphB (cut with EcoRI and PstI) and J61002 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-medium-ampicillin with colony XXX and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] J61002-RBS-tphB 65.8
aroY
- PCR on pSB1C3-aroY
- Annealing temperature: 59 °C
- Primer: RBS-aroY and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] aroY with RBS 55.2
- restriction of RBS-aroY with EcoRI and PstI
- Ligation of RBS-aroY (cut with EcoRI and PstI) and J61002 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-medium-ampicillin with colony XXX and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] J61002-RBS-aroY 77.2
Over expression
tphA1
- PCR on pSB1C3-tphA1
- Annealing temperature: 59 °C
- Primer: EcoRIGFxa-tphA1 and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphA1_over-ex 116.2
- restriction of tphA1_over-ex with EcoRI and PstI
- Ligation of tphA1_over-ex (cut with EcoRI and PstI) and pPR-IBA2 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-medium-ampicillin with colony XXX and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] pPR-IBA2-tphA1_over-ex 98.5
tphA2
- PCR on pSB1C3-tphA2
- Annealing temperature: 59 °C
- Primer: EcoRIGFxa-tphA2 and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphA2_over-ex 63.9
- restriction of tphA2_over-ex with EcoRI and PstI
- Ligation of tphA2_over-ex (cut with EcoRI and PstI) and pPR-IBA2 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-medium-ampicillin with colony XXX and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] pPR-IBA2-tphA2_over-ex 85.2
tphA3
- PCR on pSB1C3-tphA3
- Annealing temperature: 59 °C
- Primer: EcoRIGFxa-tphA3 and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphA3_over-ex 90.4
- restriction of tphA3_over-ex with EcoRI and PstI
- Ligation of tphA3_over-ex (cut with EcoRI and PstI) and pPR-IBA2 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-medium-ampicillin with colony XXX and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] pPR-IBA2-tphA3_over-ex 85.9
tphB
- PCR on pSB1C3-tphB
- Annealing temperature: 59 °C
- Primer: EcoRIGFxa-tphB and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] tphB_over-ex 87.5
- restriction of tphB_over-ex with EcoRI and PstI
- Ligation of tphB_over-ex (cut with EcoRI and PstI) and pPR-IBA2 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-medium-ampicillin with colony XXX and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] pPR-IBA2-tphB_over-ex 85.2
aroY
- PCR on pSB1C3-aroY
- Annealing temperature: 59 °C
- Primer: EcoRIGFxa-aroY and Suffix
- Both PCR products were purified via gel extraction
- Concentrations measured by Nanodrop
PCR product Concentration [ng/µl] aroY_over-ex 105.1
- restriction of aroY_over-ex with EcoRI and PstI
- Ligation of aroY_over-ex (cut with EcoRI and PstI) and pPR-IBA2 (cut with EcoRI and PstI)
- Transformation of ligation mix
- colony-PCR for verification of the transformation
- The PCR was positive
- Inoculation of 10 mL of LB-medium-ampicillin with colony XXX and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] pPR-IBA2-aroY_over-ex 92.1
week 17 (03.-07.09.12)
Operon construction
RBS-tphA1-RBS-tphA2
- restriction digest of J61002-RBS-tphA1 by EcoRI and SpeI
- Purification of insert RBS-tphA1 RBS-tphA1 (cut with EcoRI and SpeI) via gel extraction
- Concentrations measured by Nanodrop
Insert Concentration [ng/µl] RBS-tphA1 (cut with EcoRI and SpeI) 50.2
- restriction of J61002-RBS-tphA2 EcoRI and XbaI
- Dephosphorylation of the plasmid backbone J61002-RBS-tphA2 (cut with EcoRI and XbaI)
- Ligation of the plasmid backbone J61002-RBS-tphA2 (cut with EcoRI and XbaI)and RBS-tphA1 (cut with EcoRI and SpeI)
- Transformation of the ligation mix
- colony-PCR of the transformation for verification
- The PCR was positive
- Inoculation of 10 mL of LB-medium-ampicillin with colony 4 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] J61002-RBS-tphA1-RBS-tphA2 112.5
RBS-tphA3-RBS-tphB
- restriction digest of J61002-RBS-tphA3 by EcoRI and SpeI
- Purification of insert RBS-tphA3 (cut with EcoRI and SpeI) via gel extraction
- Concentrations measured by Nanodrop
Insert Concentration [ng/µl] RBS-tphA3 (cut with EcoRI and SpeI) 178.9
- restriction of J61002-RBS-tphB EcoRI and XbaI
- Dephosphorylation of the plasmid backbone J61002-RBS-tphB (cut with EcoRI and XbaI)
- Ligation of the plasmid backbone J61002-RBS-tphB (cut with EcoRI and XbaI)and RBS-tphA3 (cut with EcoRI and SpeI)
- Transformation of the ligation mix
- colony-PCR of the transformation for verification
- The PCR was positive
- Inoculation of 10 mL of LB-medium-ampicillin with colony XXX and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] J61002-RBS-tphA3-RBS-tphB 225.5
RBS-tphA1-RBS-tphA2-RBS-tphA3-RBS-tphB
- restriction of J61002-RBS-tphA1-RBS-tphA2 by EcoRI and SpeI
- Purification of insert RBS-tphA1-RBS-tphA2 (cut with EcoRI and SpeI) via gel extraction
- Concentrations measured by Nanodrop
Insert Concentration [ng/µl] RBS-tphA1-RBS-tphA2 (cut with EcoRI and SpeI) 129.5
- restriction of J61002-RBS-tphA3-RBS-tphB EcoRI and XbaI
- Dephosphorylation of the plasmid backbone J61002-RBS-tphA3-RBS-tphB (cut with EcoRI and XbaI)
- Ligation of the plasmid backbone J61002-RBS-tphA3-RBS-tphB EcoRI (cut with EcoRI and XbaI)and RBS-tphA1-RBS-tphA2 (cut with EcoRI and SpeI)
- Transformation of the ligation mix
- colony-PCR of the transformation for verification
- The PCR was positive
- Inoculation of 10 mL of LB-medium-ampicillin with colony 2 and incubation
- Miniprep of the culture and a glycerine stock was made
- Concentrations measured by Nanodrop
Miniprep Concentration [ng/µl] J61002-RBS-tphA1-RBS-tphA2-RBS-tphA3-RBS-tphB 312.2
Overexpression
tphA2
- Inoculate 10 mL of LB-medium-ampicillin with pPR-IBA2-tphA1_over-ex and incubate
- Over expression according to standard protocol
tphB
- Inoculate 10 mL of LB-medium-ampicillin with pPR-IBA2-tphA1_over-ex and incubate
- Over expression according to standard protocol
aroY
- Inoculate 10 mL of LB-medium-ampicillin with pPR-IBA2-tphA1_over-ex and incubate
- Over expression according to standard protocol
tphA1
- Inoculate 10 mL of LB-medium-ampicillin with pPR-IBA2-tphA1_over-ex and incubate
- Over expression according to standard protocol
tphA3
- Inoculate 10 mL of LB-medium-ampicillin with pPR-IBA2-tphA1_over-ex and incubate
- Over expression according to standard protocol
SDS-PAGE of tphB overexpression and tphA2 overexpression respectively
- SDS-Page according to standard protocol
| Band | Sample | Time [h] |
|---|---|---|
| 1 | tphB | 0 |
| 2 | tphB | 1 |
| 3 | tphB | 2 |
| 4 | tphB | 3 |
| 5 | tphA2 | 0 |
| 6 | tphA2 | 1 |
| 7 | tphA2 | 2 |
| 8 | tphA2 | 3 |
| 9 | Protein marker | - |
SDS-PAGE of overexpression from all five genes
- SDS-Page according to standard protocol
| Band | Sample | Time [h] |
|---|---|---|
| 1 | aroY | 0 |
| 2 | aroY | 3 |
| 3 | tphB | 0 |
| 4 | tphB | 3 |
| 5 | Protein marker | - |
| 6 | tphA1 | 0 |
| 7 | tphA1 | 3 |
| 8 | tphA2 | 0 |
| 9 | tphA2 | 3 |
| 10 | tphA3 | 0 |
| 11 | tphA3 | 3 |
| 12 | Protein marker | - |
week 18 (10.-17.09.12)
Purification of aroY
- Protein purification according to standard strep-tag purification protocol
| Band | Sample | Fraction |
|---|---|---|
| 1 | aroY | 1 |
| 2 | aroY | 2 |
| 3 | aroY | 3 and 4 together |
| 4 | aroY | 5 and 6 together |
| 5 | Protein marker | - |
Purification of TphA3
- Protein purification according to standard strep-tag purification protocol
| Band | Sample | Fraction |
|---|---|---|
| 1 | Protein marker | |
| 2 | TphA3 | Cell suspension |
| 3 | TphA3 | Cytoplasm |
| 4 | TphA3 | 1 |
| 5 | TphA3 | 2 |
| 6 | TphA3 | 3 |
| 7 | TphA3 | 4 |
| 8 | TphA3 | 5 |
| 9 | TphA3 | 6 |
| 10 | TphA3 | 7 |
| 11 | Protein marker | - |
Purification of TphA1
- Protein purification according to standard strep-tag purification protocol
| Band | Sample | Fraction |
|---|---|---|
| 1 | Protein marker | |
| 2 | TphA1 | 1 |
| 3 | TphA1 | 2 |
| 4 | TphA1 | 3 |
| 5 | TphA1 | 4 |
| 6 | TphA1 | 5 |
| 7 | TphA1 | 6 |
- Note: The other tied over TphA1 represent a contamination of aroY
week 19 (17.-21.09.12)
tphA1
tphA2
tphA3
tphB
aroY
Other
 "
"