Team:Stanford-Brown/HellCell/pH
pH: Base
At a glance
Extremophile: Escherichia coli
Proteins of interest: serine deaminase (serine catabolism)
Consensus: Effective
Alkaliphiles like marine bacteria (pH 8.2) and pyloric duct flora (pH ~10 or higher) rely largely on transmembrane transporter proteins to regulate the pH within the cytoplasm and thrive (Padan, Bibi, Ito, and Krulwich 2005). Most notable of these transporters are ATP synthase and cation/proton antiporters. Both of these use some type of energy from the cell to power the transport of H+ ions across the membrane. In the former, the energy released in the dephosphorylation of ATP is coupled with the transport; in the latter, an electrochemical gradient is exploited (Padan, et al. 2005).
Unfortunately, there are several issues with using this direct method. Just the structural genes (not considering regulation) required for these transmembrane proteins total ~10 kb in length, which is difficult to manage in BioBrick form. Additionally, these proteins are optimized for the membranes of alkaliphilic bacteria which have a different lipid composition than E. coli, making their behavior tricky to control in a planned fashion (Padan, et al. 2005). Finally, the actions of both can be reversed by a strong proton gradient (after all, ATP synthase is used to synthesize ATP from a H+ gradient), requiring much more extensive investigation of regulation mechanisms.
However, the Hell Cell squad found it intriguing that E. coli increases the catabolism of amino acids when exposed to high pHs, most researched of which are tryptophan and serine (Padan et al.). This process creates buffers in the cytoplasm to help counter the effects of the high pH. Since the heightened catabolism of tryptophan would require the insertion of a tryptophan transporter, we chose serine catabolism and isolated the gene for serine deaminase from E. coli.
We inserted this gene into the Test Plasmid, transformed E. coli with it, and assayed its function. We also tested the effectiveness of two genes from our radiation assays, Dps and recA, as DNA repair mechanisms also are helpful in differing pH conditions--see Radiation.
Assay
Liquid cultures of negative control, recA, dps, and sdaB transformed NEB5α E. coli were grown up over night at 37°C. After incubation, LB + amp media of pH 3.54, 4.60, 6.93, 9.07, 9.50 were created using HCl and NaOH. In a 96 well plate, 8 wells were filled with LB + amp blank. Next, 5uL of each transformed bacteria was placed in five different pH wells. This was replicated four times, thus, creating a 96 well plate with 8 blanks, and four replicates of each test construct at five different pH’s. Finally, the OD600 was measured every five minutes for 24 hours while the plate was incubated at 37°C, shook for 1 minute before every sample and between every sample in a spectrophotometer.
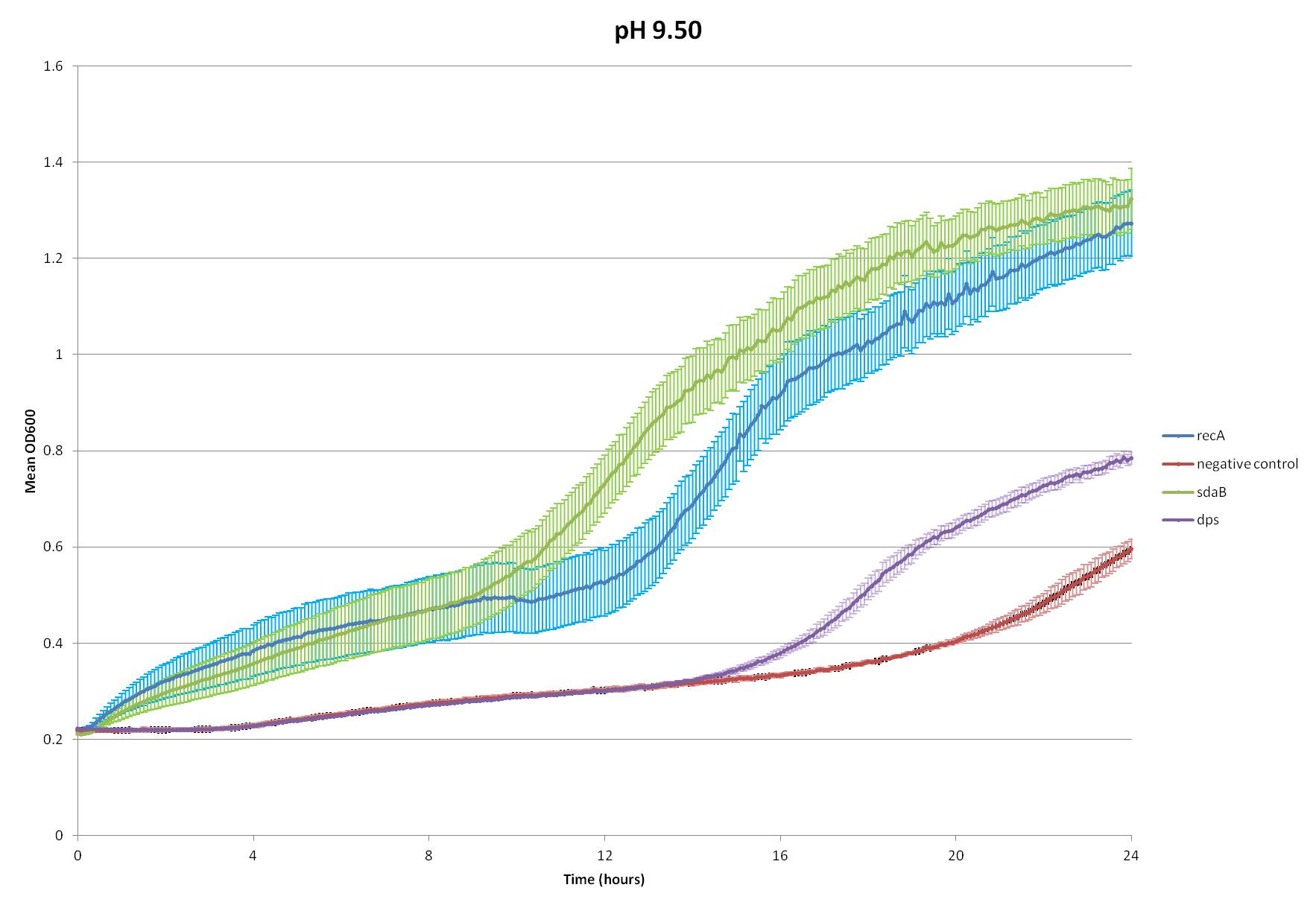
Figure 1: This graph displays the optical density at 600nm and standard error of the mean between four replicates of four different constructs. Optical density was measured over a 24 hours period every five minutes. The cultures where incubated at 37°C and shook for 1 minute before every five minute period and for 1 minute during every five minute period.
Conclusions
Based on Figure 1, it can be clearly seen that there are two constructs which clearly provide resistance to basic conditions and one construct which may provide resistance to basic conditions but to a lesser extent. SdaB and recA both dramatically outgrow the negative control. Additionally, E. coli transformed with Dps seem to show slightly better growth performance than negative control. Since it is clear that E.coli transformed with sdaB and recA reach a higher optical density in a shorter period of time, it is reasonable to assert that these two genes provide base resistance for pH’s up to 9.5. Furthermore, it is also possible that they may help at higher pH’s, and it might be worthwhile to study the effects of higher pH's in future assays. Nonetheless, both sdaB and recA clearly display their ability to increase resistance to basic conditions.
Why not Acid??
So, why didn’t we include acid, you ask? We did not forget it, we promise. Like alkaliphiles, acidophiles use ATP synthases to maintain their internal pH at hospitable levels. However, as already mentioned, these synthases are large and difficult to control (Foster 2004). Acidophiles also have membranes made up of tetraether lipids instead of ester linkages, which is difficult to engineer (Baker-Austin and Dopson 2007).
Helicobacter pylori, a bacterium that inhabits the stomach and causes ulcers, thrives in the corrosively acidic environment of the stomach (pH 2-5). Central to this capacity is the breakdown of urea into ammonia and carbon dioxide, catalyzed by the enzyme urease (Labigne, Cussac, and Courcoux 1991). It was found, however, that several genes are necessary for expression of urease and urease-like phenotypes in E. coli, making it difficult to BioBrick (Cussac, Ferrero, Labigne 1992). The 2011 Brown-Stanford iGEM team made significant headway in isolating and characterizing urease from Sporosarcina pasteurii, but because they were not able to obtain the sequence of this gene, we did not decide to pursue it.
Amino acid degradation is another mechanism common to both alkaliphiles and acidophiles—both glutamate and arginine can be broken down to produce buffers (Foster 2004). Arginine, however, is not present in high concentrations in LB broth, and the three genes involved in glutamate catabolism (gadA, gadB, and gadC) had several BioBrick restriction enzyme cut sites which we did not have time to engineer around.
Sources:
Padan, E., Bibi, E., Ito, M., Krulwich, T.A. (2005). Alkaline pH Homeostasis in Bacteria: New Insights. Biochim Biophys Acta, 1717(2): 67-88.
Foster, J. W. (2004). Escherichia Coli acid resistance: Tales of an amateur acidophile. Nature Rev. Microbiol., 2, 898-907.
Labigne, A., Cussac, V., Courcoux, P. (1991). Shuttle cloning and nucleotide sequences of Helicobacter pylori genes responsible for urease activity. J. Bacteriol., 173(6): 1920-1931.
Cussac, V., Ferrero, R. L., Labigne, A. (1992). Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J. Bacteriol., 174(8): 2466-2473.
Baker-Austin, C. Dopson, M. (2007). Life in acid: pH homeostasis in acidophiles. Trends in Microbiology, 15(4): 165-170.
 "
"