Team:Stanford-Brown/HellCell/Cold
From 2012.igem.org
Vishesh.jain (Talk | contribs) |
Vishesh.jain (Talk | contribs) |
||
| (13 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
<li><a href="/Team:Stanford-Brown/HellCell/Introduction" id="project">Hell Cell:</a></li> | <li><a href="/Team:Stanford-Brown/HellCell/Introduction" id="project">Hell Cell:</a></li> | ||
<li><a href="/Team:Stanford-Brown/HellCell/Introduction">Introduction</a></li> | <li><a href="/Team:Stanford-Brown/HellCell/Introduction">Introduction</a></li> | ||
| + | <li><a href="/Team:Stanford-Brown/HellCell/Plasmid">Test Plasmid</a></li> | ||
<li id="active"><a href="#" id="current">Cold</a></li> | <li id="active"><a href="#" id="current">Cold</a></li> | ||
<li><a href="/Team:Stanford-Brown/HellCell/Desiccation"> Desiccation </a></li> | <li><a href="/Team:Stanford-Brown/HellCell/Desiccation"> Desiccation </a></li> | ||
<li><a href="/Team:Stanford-Brown/HellCell/Radiation">Radiation</a></li> | <li><a href="/Team:Stanford-Brown/HellCell/Radiation">Radiation</a></li> | ||
<li><a href="/Team:Stanford-Brown/HellCell/pH">pH</a></li> | <li><a href="/Team:Stanford-Brown/HellCell/pH">pH</a></li> | ||
| - | <li><a href="/Team:Stanford-Brown/ | + | <li><a href="/Team:Stanford-Brown/Parts">BioBricks</a></li> |
| + | <li><a href="https://docs.google.com/document/d/1Pe9voM2l_nrVJk0hwzJ6z6tCCMn9ckykVNX5nLZ8qGg/edit">Lab Notebook</a></li> | ||
| + | <li><a href="/Team:Stanford-Brown/Protocols">Protocols</a></li> | ||
</ul> | </ul> | ||
</div> | </div> | ||
| Line 18: | Line 21: | ||
== '''Cold''' <br> <br> <font size = 2.5> <u> At a glance</u> <br> Extremophile: ''Psychromonas ingrahamii'' <br> Proteins of interest: choline dehydrogenase and betaine aldehyde dehydrogenase (glycine betaine biosynthesis pathway) <br> Consensus: Inconclusive </font> == | == '''Cold''' <br> <br> <font size = 2.5> <u> At a glance</u> <br> Extremophile: ''Psychromonas ingrahamii'' <br> Proteins of interest: choline dehydrogenase and betaine aldehyde dehydrogenase (glycine betaine biosynthesis pathway) <br> Consensus: Inconclusive </font> == | ||
| - | There are a few | + | There are a few common mechanisms that bacteria use to survive at low temperatures that are well-characterized. The most important two of these are (1) producing enzymes that have specific adaptations that allow them to function optimally at lower temperatures and (2) incorporating fatty acids and carotenoids in their membranes to make them more fluid and less freeze-prone (Chattopadhyay 2006). More recent research has drawn attention to certain bacteria’s ability to use different metabolic pathways to conserve ATP, upregulate the production of cold shock proteins, and keep dividing exponentially at surprisingly low temperatures (Chattopadhyay 2006). However, transferring these mechanisms to a bacterium normally not cold-tolerant requires an almost complete overhaul of the bacterium’s metabolism, which is slow to respond and is difficult to BioBrick due to the lengths of the genes involved. Such engineering could also potentially cause the bacterium to lose its ability to thrive under normal conditions. |
| - | More approachable methods of engineering cold-tolerant bacteria are to make them produce cryprotectants or antifreeze proteins (Chattopadhyay 2006). The Yale 2011 iGEM team made significant progress in antifreeze proteins, developing an antifreeze protein optimized for ''E. coli'' based on that of the cold-tolerant ribbed pine borer beetle (''Rhagium iniquisitor''), which survives on pines in Siberia. The Hell Cell squad therefore decided to explore the realm of cryoprotectants, focusing on the glycine betaine pathway found in bacteria. | + | More approachable methods of engineering cold-tolerant bacteria are to make them produce cryprotectants or antifreeze proteins (Chattopadhyay 2006). The Yale 2011 iGEM team made significant progress in antifreeze proteins, developing an antifreeze protein optimized for ''E. coli'' based on that of the cold-tolerant ribbed pine borer beetle (''Rhagium iniquisitor''), which survives on pines in Siberia. The Hell Cell squad therefore decided to explore the realm of cryoprotectants, focusing on the glycine betaine pathway naturally found in many bacteria. |
| - | Glycine betaine is a chemical that prevents the aggregation of proteins at low temperatures and facilitates the process of maintaining membrane fluidity (Chattopadhyay 2006). ''Psychromonas ingrahamii'', an especially cold-tolerant bacteria, grows exponentially at -12° C, and it is believed that the production of glycine betaine from choline is essential for it to maintain osmotic stability at this temperature. A pathway to produce glycine betaine also natively exists in ''E. coli'', but | + | Glycine betaine is a chemical that prevents the aggregation of proteins at low temperatures and facilitates the process of maintaining membrane fluidity (Chattopadhyay 2006). ''Psychromonas ingrahamii'', an especially cold-tolerant bacteria, grows exponentially at -12° C, and it is believed that the production of glycine betaine from choline is essential for it to maintain osmotic stability at this temperature. A pathway to produce glycine betaine also natively exists in ''E. coli'', but is not upregulated enough to afford it the cold resistance of ''P. ingrahamii''. |
| - | Since we did not have access to ''P. ingrahamii'', we isolated the pathway from ''E. coli'' (the genes of which have an ~86% to those of ''P. ingrahamii'' when compared using MegaBlast) and upregulated its production using | + | Since we did not have access to ''P. ingrahamii'', we isolated the pathway from ''E. coli'' (the genes of which have an ~86% match to those of ''P. ingrahamii'' when compared using MegaBlast) and upregulated its production using our Test Plasmid strategy (see Test Plasmid). The proteins of interest have identical function in the two organisms and we placed them under a new promoter, so genetic differences between ''E. coli'' and ''P. ingrahamii'' are largely irrelevant in this context. |
| - | After transforming bacteria with our test plasmid, we grew approximately equal amounts of liquid culture of the negative control (see | + | '''Assay''' |
| + | |||
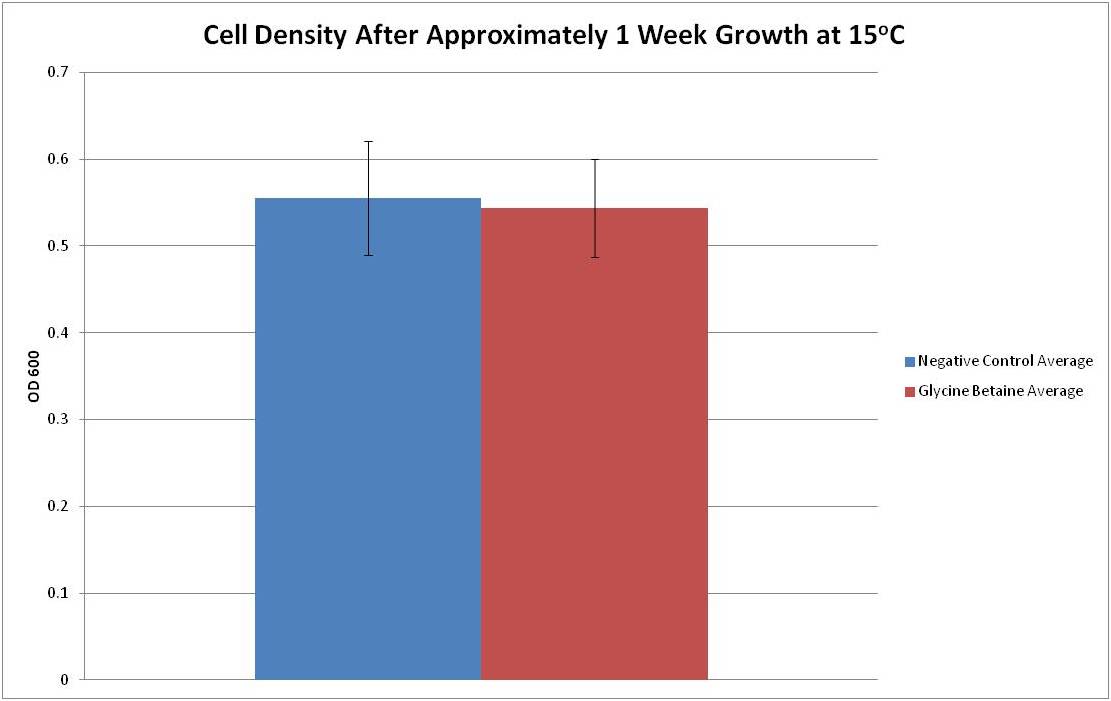
| + | After transforming bacteria with our test plasmid, we grew approximately equal amounts of liquid culture of the negative control and betaine transformant(see Test Plasmid) in LB+amp at 15°C, the lower limit of ''E. coli''’s survival range. OD measurements were taken at time points t = 0 days and t = 9 days. The data is presented below. | ||
[[File:Cold_graph.jpeg|800px|center]] | [[File:Cold_graph.jpeg|800px|center]] | ||
| + | Figure 1: The average OD600 and standard deviation of six cultures grown for one week at 15oC. The negative control is represented in blue and the glycine betaine transformant is represented in red. | ||
| + | |||
| + | '''Conclusions''' | ||
| + | |||
| + | From Figure 1, it can be seen that the average optical density at 600nm after 1 week growth at 15°C for both negative control and glycine betaine transformed bacteria is between 0.5 and 0.6. This data seems to suggest that the growth rates for the tested growth condition do not differ between the negative control and our transformants. However, it is possible that lower temperatures are required to obtain a result where the construct boosts cold resistance significantly. Ideally, a lower temperature where the negative control has very limited growth would have been assayed; however, due to time constraints and slower growth rates at lower temperatures, such testing was not possible. Further tests are required in order to fully characterize the glycine betaine construct’s ability to provide cold resistance. | ||
| - | |||
Sources: | Sources: | ||
| + | |||
Chattopadhyay, M. K. (2006). Mechanism of bacterial adaptation to low temperature. ''J. Biosci., 31'' (1), 157-165. | Chattopadhyay, M. K. (2006). Mechanism of bacterial adaptation to low temperature. ''J. Biosci., 31'' (1), 157-165. | ||
Riley, M., Staley, J. T., Danchin, A., Wang, T. Z., Brettin, T. S., Hauser, L. J., Land, M. L., Thompson, L. S. (2008). Genomics of an extreme psychrophile, ''Psychromonas ingrahamii''. ''BMC Genomics, 9''(210), 1-19. | Riley, M., Staley, J. T., Danchin, A., Wang, T. Z., Brettin, T. S., Hauser, L. J., Land, M. L., Thompson, L. S. (2008). Genomics of an extreme psychrophile, ''Psychromonas ingrahamii''. ''BMC Genomics, 9''(210), 1-19. | ||
Latest revision as of 02:05, 4 October 2012
Cold
At a glance
Extremophile: Psychromonas ingrahamii
Proteins of interest: choline dehydrogenase and betaine aldehyde dehydrogenase (glycine betaine biosynthesis pathway)
Consensus: Inconclusive
There are a few common mechanisms that bacteria use to survive at low temperatures that are well-characterized. The most important two of these are (1) producing enzymes that have specific adaptations that allow them to function optimally at lower temperatures and (2) incorporating fatty acids and carotenoids in their membranes to make them more fluid and less freeze-prone (Chattopadhyay 2006). More recent research has drawn attention to certain bacteria’s ability to use different metabolic pathways to conserve ATP, upregulate the production of cold shock proteins, and keep dividing exponentially at surprisingly low temperatures (Chattopadhyay 2006). However, transferring these mechanisms to a bacterium normally not cold-tolerant requires an almost complete overhaul of the bacterium’s metabolism, which is slow to respond and is difficult to BioBrick due to the lengths of the genes involved. Such engineering could also potentially cause the bacterium to lose its ability to thrive under normal conditions.
More approachable methods of engineering cold-tolerant bacteria are to make them produce cryprotectants or antifreeze proteins (Chattopadhyay 2006). The Yale 2011 iGEM team made significant progress in antifreeze proteins, developing an antifreeze protein optimized for E. coli based on that of the cold-tolerant ribbed pine borer beetle (Rhagium iniquisitor), which survives on pines in Siberia. The Hell Cell squad therefore decided to explore the realm of cryoprotectants, focusing on the glycine betaine pathway naturally found in many bacteria.
Glycine betaine is a chemical that prevents the aggregation of proteins at low temperatures and facilitates the process of maintaining membrane fluidity (Chattopadhyay 2006). Psychromonas ingrahamii, an especially cold-tolerant bacteria, grows exponentially at -12° C, and it is believed that the production of glycine betaine from choline is essential for it to maintain osmotic stability at this temperature. A pathway to produce glycine betaine also natively exists in E. coli, but is not upregulated enough to afford it the cold resistance of P. ingrahamii.
Since we did not have access to P. ingrahamii, we isolated the pathway from E. coli (the genes of which have an ~86% match to those of P. ingrahamii when compared using MegaBlast) and upregulated its production using our Test Plasmid strategy (see Test Plasmid). The proteins of interest have identical function in the two organisms and we placed them under a new promoter, so genetic differences between E. coli and P. ingrahamii are largely irrelevant in this context.
Assay
After transforming bacteria with our test plasmid, we grew approximately equal amounts of liquid culture of the negative control and betaine transformant(see Test Plasmid) in LB+amp at 15°C, the lower limit of E. coli’s survival range. OD measurements were taken at time points t = 0 days and t = 9 days. The data is presented below.
Figure 1: The average OD600 and standard deviation of six cultures grown for one week at 15oC. The negative control is represented in blue and the glycine betaine transformant is represented in red.
Conclusions
From Figure 1, it can be seen that the average optical density at 600nm after 1 week growth at 15°C for both negative control and glycine betaine transformed bacteria is between 0.5 and 0.6. This data seems to suggest that the growth rates for the tested growth condition do not differ between the negative control and our transformants. However, it is possible that lower temperatures are required to obtain a result where the construct boosts cold resistance significantly. Ideally, a lower temperature where the negative control has very limited growth would have been assayed; however, due to time constraints and slower growth rates at lower temperatures, such testing was not possible. Further tests are required in order to fully characterize the glycine betaine construct’s ability to provide cold resistance.
Sources:
Chattopadhyay, M. K. (2006). Mechanism of bacterial adaptation to low temperature. J. Biosci., 31 (1), 157-165.
Riley, M., Staley, J. T., Danchin, A., Wang, T. Z., Brettin, T. S., Hauser, L. J., Land, M. L., Thompson, L. S. (2008). Genomics of an extreme psychrophile, Psychromonas ingrahamii. BMC Genomics, 9(210), 1-19.
 "
"