Team:SEU O China/Notebook
From 2012.igem.org
| Line 77: | Line 77: | ||
| - | PCR/Gel Extraction | + | '''PCR/Gel Extraction''' |
| - | + | ||
| - | + | Fresh TAE buffer as running buffer is recommended. Reusing running buffer will result the increase of the pH and then reduce yields. | |
| - | + | ||
| - | + | #For cycle-pure (PCR reaction): Add 2 volumes of Buffer GC to 1 volume of the PCR reaction and mix completely by vortexing. Briefly spin the tube to collect any drops from the inside wall and tube lid. For PCR products less than 200 bp, add 5 volumes of Buffer GC to 1 volume of PCR reaction | |
| - | + | #For agarose gel: Excise the DNA fragment from the agarose gel and weigh it in a 1.5 mL microtube. Add 1 volume of Buffer GC to 1 volume of gel to the 1.5 mL microtube and incubate the mixture at 55-60°C for 8 min. Mix the tube by tapping the bottom every 2 min till the gel has melted completely. Cool the tube to room temperature.'' Note: A gel slice of 100 mg approximately equals to 100 μL. For DNA fragment less than 200 bp, add 1 volume of isopropanol.'' | |
| - | + | #Transfer up to 700 μL DNA/Buffer GC mixture to a spin column with a collection tube. Centrifuge at 13,000 x g for 1 min at room temperature. Discard the flow-through and put the column back to the collection tube. Repeat this step to process the remaining solution. | |
| - | Note: Ensure that ethanol has been added to DNA Wash Buffer as instructed . | + | #Add 300 μL of Buffer GC into the DNA Mini Column. Centrifuge at 13,000 x g for 30 to 60 seconds at room temperature to wash the DNA Mini Column. |
| - | + | #Add 650 μL DNA Wash Buffer to the column and centrifuge at 13,000 x g for 30 s at room temperature. Discard the flow through and insert the column, with the lid open, back to the collection tube. Repeat step “4”. ''Note: Ensure that ethanol has been added to DNA Wash Buffer as instructed .'' | |
| - | Note: The residual ethanol will be removed more efficiently with the column lid open during centrifugation. | + | #Centrifuge the empty DNA column, with the lid open, at 13,000 x g for 2 min to dry the ethanol residue in the matrix. ''Note: The residual ethanol will be removed more efficiently with the column lid open during centrifugation.'' |
| - | + | #Place the column into a clean 1.5 mL micocentrifuge tube and add 30-50 μL pre-warmed (60oC) Elution Buffer or ddH2O to the center of the column. Incubate at room temperature for 1 min. Centrifuge at 13,000 x g for 1 min | |
| + | |||
| + | |||
| + | |||
Miniprep Plasmids | Miniprep Plasmids | ||
| - | + | #Inoculate 1-4 mL LB containing appropriate antibiotic with a fresh colony from a freshly streaked selective plate. Incubate at 37°C for 14-16 hours with vigorous shaking. | |
| - | Note: Prolonged incubation (> 16 hours) is not recommended since the E.coli starts to lyse and the plasmid yields may be reduced. | + | ''Note: Prolonged incubation (> 16 hours) is not recommended since the E.coli starts to lyse and the plasmid yields may be reduced. Do not grow the culture directly from the glycerol stock. This protocol is optimized for E. coli strain cultured in LB medium. When using TB or 2xYT medium, special care needs to be taken to ensure the cell density doesn’t exceed 3.0 (OD600). Buffers need to be scaled up proportionally if over amount of cultures are being processed.'' |
| - | + | #Harvest the bacterial culture by centrifugation for 1 min at 10,000 rpm. Pour off the supernatant and blot the inverted tube on a paper towel to remove residue medium. Remove the residue medium completely. ''Note: Residue medium will cause,'' | |
| - | Note: Residue medium will cause, | + | #*Poor cell lysis and thus lower DNA yield. |
| - | + | #*Loose pellet after centrifugation in step 6. | |
| - | + | #Add 250 μL Buffer A1 (Add RNase A to Buffer A1 before use) and completely resuspend bacterial pellet by vortexing or pipetting. | |
| - | + | ''Note: Complete resuspension is critical for bacterial lysis and lysate neutralization.'' | |
| - | Note: Complete resuspension is critical for bacterial lysis and lysate neutralization. | + | #Add 250 μL Buffer B1, mix gently by inverting the tube 10 times (do not vortex), and incubate at room temperature for 5 minutes. |
| - | + | ''Note: Do not incubate for more than 5 minutes. Note: Buffer B1 precipitates (cloudy look) below room temperature. Warm up Buffer B1 at 50°C to dissolve precipitation before use.'' | |
| - | Note: Do not incubate for more than 5 minutes. Note: Buffer B1 precipitates (cloudy look) below room temperature. Warm up Buffer B1 at 50°C to dissolve precipitation before use. | + | #Add 350 μL Buffer N1, mix completely by inverting/shaking the vial for 5 times and sharp hand shaking for 2 times. |
| - | + | ''Note: Incubating the lysate in ice for 1 min will improve the yield. Note: It is critical to mix the solution well. If the mixture still appears conglobated, brownish or viscous, more mixing is required to completely neutralize the solution.'' | |
| - | Note: Incubating the lysate in ice for 1 min will improve the yield. Note: It is critical to mix the solution well. If the mixture still appears conglobated, brownish or viscous, more mixing is required to completely neutralize the solution. | + | #Centrifuge the lysate at 13,000 rpm for 10 minutes at room temperature. |
| - | + | ''Note: If the lysate doesn’t appear clean, reverse the tube angle, centrifuge for 5 more minutes and then transfer the clear lysate to DNA column. '' | |
| - | Note: If the lysate doesn’t appear clean, reverse the tube angle, centrifuge for 5 more minutes and then transfer the clear lysate to DNA column. | + | #Carefully transfer the clear lysate into a DNA column with a collection tube, avoid the precipitations, spin at 13,000 rpm for 1 minute, discard the flow-through and put the column back to the collection tube. |
| - | + | #Optional: Add 500 μL Buffer KB into the spin column, centrifuge at 13,000 rpm for 1 minute. Remove the spin column from the tube and discard the flow-through. Put the column back to the collection tube. | |
| - | + | ''Note: Buffer KB is recommended for endA+ strains such as HB101, JM101, TG1 or their derived strains. It is not necessary for isolating DNA from endA- strains such as Top 10 and DH5a.'' | |
| - | Note: Buffer KB is recommended for endA+ strains such as HB101, JM101, TG1 or their derived strains. It is not necessary for isolating DNA from endA- strains such as Top 10 and DH5a. | + | #Add 650 μL DNA Wash Buffer (Add ethanol to DNA wash buffer before use) into the spin column, centrifuge at 13,000 rpm for 1 minute at room temperature. Remove the spin column from the tube and discard the flow-through. Repeat step “9” to improve the recovery. |
| - | + | #Reinsert the spin column, with the lid open, into the collection tube and centrifuge for 2 minutes at 13,000 rpm. | |
| - | + | ''Note: Residual ethanol can be removed more efficiently with the column lid open. It is critical to remove residual ethanol completely.'' | |
| - | Note: Residual ethanol can be removed more efficiently with the column lid open. It is critical to remove residual ethanol completely. | + | #Carefully transfer the spin column into a sterile 1.5 mL tube and add 50-100 μL (> 50 μL) Sterile ddH20 or Elution Buffer into the center of the column and let it stand for 2 minutes. Elute the DNA by centrifugation at 13,000 rpm for 1 minute. Reload the eluate into the column and elute again. |
| - | + | ''Note: If ddH2O is applied, please make sure the pH is no less than 7.0 (7.0-8.5 is preferred). NaOH could be used to adjust the pH of ddH2O. The DNA is ready for downstream applications such ascloning/subcloning, RFLP, library screening, in vitro translation, sequencing, transfection of robust cells such as HEK293 cells. Note: It’s highly recommended to remove the endotoxin (PD1212) if the DNA is used for endotoxin-sensitive cell lines, primary cultured cells or micro-injection. '' | |
| - | Note: If ddH2O is applied, please make sure the pH is no less than 7.0 (7.0-8.5 is preferred). NaOH could be used to adjust the pH of ddH2O. | + | #The DNA concentration can be calculated as follows, Concentration (μg/mL) = OD260nm x 50 x dilution factor. |
| - | + | ||
| - | Concentration (μg/mL) = OD260nm x 50 x dilution factor | + | |
| + | |||
Transforming DNA from Distribution Plates: | Transforming DNA from Distribution Plates: | ||
1. Thaw competent cells on ice. | 1. Thaw competent cells on ice. | ||
Revision as of 09:04, 23 September 2012


Notebook
Protocols
LB Medium:
Liquid:
- 10 g of tryptone,
- 5 g of yeast extract,
- 10 g of NaCl,
- 1 liter of distilled water
Solid:
- 10 g of tryptone,
- 5 g of yeast extract,
- 10 g of NaCl,
- 15 g of agar,
- 1 liter of distilled water
Biobrick Assembly Restriction Digest:
For a double digest:
- To a pcr tube, add 15 ul of miniprepped plasmid or purified PCR product
- Add 3 ul of the appropriate buffer
- Add 1ul of each enzyme.
- Incubate at 4˚C overnight.
Biobrick Assembly Ligation:
- In a PCR tube, add restriction digested and PCR purified insert to backbone in a 4:1 molar ratio, usually 1 ul backbone with 4 ul insert works.
- Add 10ul T4 ligase buffer
- Add 1 ul T4 ligase
- Create a negative control replacing the insert with water
- Add reactions to thermal cycler, and leave at 4˚C overnight.
- Use 2 ul of the ligation reactions to transform cells.
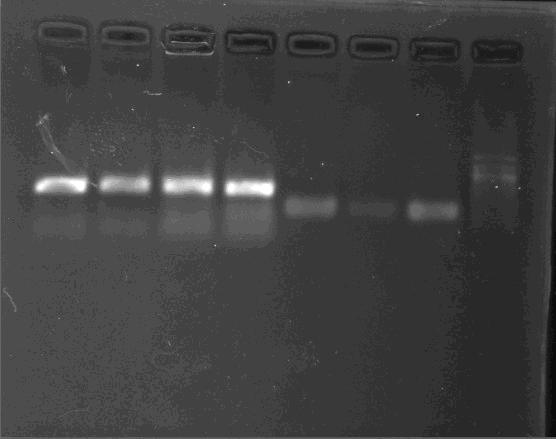
PCR/Gel Extraction
Fresh TAE buffer as running buffer is recommended. Reusing running buffer will result the increase of the pH and then reduce yields.
- For cycle-pure (PCR reaction): Add 2 volumes of Buffer GC to 1 volume of the PCR reaction and mix completely by vortexing. Briefly spin the tube to collect any drops from the inside wall and tube lid. For PCR products less than 200 bp, add 5 volumes of Buffer GC to 1 volume of PCR reaction
- For agarose gel: Excise the DNA fragment from the agarose gel and weigh it in a 1.5 mL microtube. Add 1 volume of Buffer GC to 1 volume of gel to the 1.5 mL microtube and incubate the mixture at 55-60°C for 8 min. Mix the tube by tapping the bottom every 2 min till the gel has melted completely. Cool the tube to room temperature. Note: A gel slice of 100 mg approximately equals to 100 μL. For DNA fragment less than 200 bp, add 1 volume of isopropanol.
- Transfer up to 700 μL DNA/Buffer GC mixture to a spin column with a collection tube. Centrifuge at 13,000 x g for 1 min at room temperature. Discard the flow-through and put the column back to the collection tube. Repeat this step to process the remaining solution.
- Add 300 μL of Buffer GC into the DNA Mini Column. Centrifuge at 13,000 x g for 30 to 60 seconds at room temperature to wash the DNA Mini Column.
- Add 650 μL DNA Wash Buffer to the column and centrifuge at 13,000 x g for 30 s at room temperature. Discard the flow through and insert the column, with the lid open, back to the collection tube. Repeat step “4”. Note: Ensure that ethanol has been added to DNA Wash Buffer as instructed .
- Centrifuge the empty DNA column, with the lid open, at 13,000 x g for 2 min to dry the ethanol residue in the matrix. Note: The residual ethanol will be removed more efficiently with the column lid open during centrifugation.
- Place the column into a clean 1.5 mL micocentrifuge tube and add 30-50 μL pre-warmed (60oC) Elution Buffer or ddH2O to the center of the column. Incubate at room temperature for 1 min. Centrifuge at 13,000 x g for 1 min
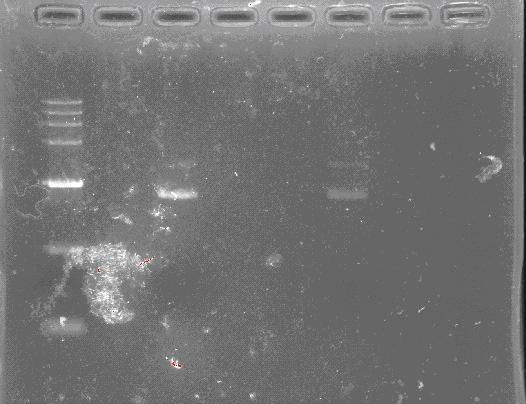
Miniprep Plasmids
- Inoculate 1-4 mL LB containing appropriate antibiotic with a fresh colony from a freshly streaked selective plate. Incubate at 37°C for 14-16 hours with vigorous shaking.
Note: Prolonged incubation (> 16 hours) is not recommended since the E.coli starts to lyse and the plasmid yields may be reduced. Do not grow the culture directly from the glycerol stock. This protocol is optimized for E. coli strain cultured in LB medium. When using TB or 2xYT medium, special care needs to be taken to ensure the cell density doesn’t exceed 3.0 (OD600). Buffers need to be scaled up proportionally if over amount of cultures are being processed.
- Harvest the bacterial culture by centrifugation for 1 min at 10,000 rpm. Pour off the supernatant and blot the inverted tube on a paper towel to remove residue medium. Remove the residue medium completely. Note: Residue medium will cause,
- Poor cell lysis and thus lower DNA yield.
- Loose pellet after centrifugation in step 6.
- Add 250 μL Buffer A1 (Add RNase A to Buffer A1 before use) and completely resuspend bacterial pellet by vortexing or pipetting.
Note: Complete resuspension is critical for bacterial lysis and lysate neutralization.
- Add 250 μL Buffer B1, mix gently by inverting the tube 10 times (do not vortex), and incubate at room temperature for 5 minutes.
Note: Do not incubate for more than 5 minutes. Note: Buffer B1 precipitates (cloudy look) below room temperature. Warm up Buffer B1 at 50°C to dissolve precipitation before use.
- Add 350 μL Buffer N1, mix completely by inverting/shaking the vial for 5 times and sharp hand shaking for 2 times.
Note: Incubating the lysate in ice for 1 min will improve the yield. Note: It is critical to mix the solution well. If the mixture still appears conglobated, brownish or viscous, more mixing is required to completely neutralize the solution.
- Centrifuge the lysate at 13,000 rpm for 10 minutes at room temperature.
Note: If the lysate doesn’t appear clean, reverse the tube angle, centrifuge for 5 more minutes and then transfer the clear lysate to DNA column.
- Carefully transfer the clear lysate into a DNA column with a collection tube, avoid the precipitations, spin at 13,000 rpm for 1 minute, discard the flow-through and put the column back to the collection tube.
- Optional: Add 500 μL Buffer KB into the spin column, centrifuge at 13,000 rpm for 1 minute. Remove the spin column from the tube and discard the flow-through. Put the column back to the collection tube.
Note: Buffer KB is recommended for endA+ strains such as HB101, JM101, TG1 or their derived strains. It is not necessary for isolating DNA from endA- strains such as Top 10 and DH5a.
- Add 650 μL DNA Wash Buffer (Add ethanol to DNA wash buffer before use) into the spin column, centrifuge at 13,000 rpm for 1 minute at room temperature. Remove the spin column from the tube and discard the flow-through. Repeat step “9” to improve the recovery.
- Reinsert the spin column, with the lid open, into the collection tube and centrifuge for 2 minutes at 13,000 rpm.
Note: Residual ethanol can be removed more efficiently with the column lid open. It is critical to remove residual ethanol completely.
- Carefully transfer the spin column into a sterile 1.5 mL tube and add 50-100 μL (> 50 μL) Sterile ddH20 or Elution Buffer into the center of the column and let it stand for 2 minutes. Elute the DNA by centrifugation at 13,000 rpm for 1 minute. Reload the eluate into the column and elute again.
Note: If ddH2O is applied, please make sure the pH is no less than 7.0 (7.0-8.5 is preferred). NaOH could be used to adjust the pH of ddH2O. The DNA is ready for downstream applications such ascloning/subcloning, RFLP, library screening, in vitro translation, sequencing, transfection of robust cells such as HEK293 cells. Note: It’s highly recommended to remove the endotoxin (PD1212) if the DNA is used for endotoxin-sensitive cell lines, primary cultured cells or micro-injection.
- The DNA concentration can be calculated as follows, Concentration (μg/mL) = OD260nm x 50 x dilution factor.
Transforming DNA from Distribution Plates: 1. Thaw competent cells on ice. 2. Add 10 microliters of pure water to each well of DNA from plates, pipette up and down. 3. Transfer into storage tube. 4. Pipette 1-2 microliters of the DNA into the competent cell tubes. 5. Stir with pipette tip, gently flick tube. 6. Leave on ice for 30 minutes. 7. Heat Shock for 45 sec by using a water bath set to 42°C and then chill on ice for 2 min. 8. Pipette 500 micro Liters of S.O.C. (LB + glucose) into 14ml culture tubes; transfer the competent cells into these tubes and incubate in a 37 degree shaker for 0-60 minutes before plating. 9. For source plate DNA, plate 100 microliters. Taq PCR Set up tubes with 7 ul H20, 1 ul template, 1 ul forward primer, 1 ul reverse primer, 10 ul PCR mix Initial denaturation: 94°C for 1:30 min 35 cycles of: 94°C for 0:30 min 54°C for 1:00 min 72°C for 1:00 min/Kb Final extension: 72°C for 6:00 min Colony PCR Suspend colonies in 5 ul H20 Lyse with 98°C incubation for 10 minutes Use 1 ul of this suspension as template Set up tubes with 7 ul H20, 1 ul template, 1 ul forward primer, 1 ul reverse primer, 10 ul PCR mix Cycle: 2 minutes at 98°C Run 30 cycles of: 10 seconds at 98°C 30 seconds at 55°C-65°C 40 seconds at 72°C Final extension: 5 minutes at 72°C
March
2012-3-18 Sun
After discussion in the seminar, 13 students organized the iGEM team and set out to design our project.
2012-3-24 Sat
Our project application were admitted in our school and the registry work started.
The achievements of one week's work:
Muyuan Chen & Xuelong Fan: responsible for the fund application and team registry work.
Xingyu Zhou: designed the bio-sensor based on the asymmetric colony.
Ying Cheng & Zhu Zhu: Searched the information about quorum sensing.
Xiaodan Xu: Read the literature on E.coli's flagella and division.
Peng Cheng & Le Zhou: designed the wiki and logo.
2012-3-25 Sun
Shen Gao: improved the method of cultivation of E.coli.
2012-3-26 Tue
Muyuan Chen: Did some research on light sensor. We voted and named our team SEU_Omega~
2012-3-27 Wed
Filled the application form on the www.igem.org.
2012-3-31 Sat
Xintong Ling: Finished the simulation of diffusion equation in the rectangular case.
April
2012-4-1 Sun
Xintong Ling: finished the diffusion model based on the random containers and made a plan for further models.
2012-4-5 Thur
Muyuan Chen: Made the schedule and experiment program before our summer vacation. Found some information about toggle switch.
Xuelong Fan: Did some research on the light sensor.
Xiaodan Xu: Learnt about how to use NCBI.
2012-4-7 Sat
Had a group meeting and discussed about the future plans.
Xiaodan Xu: Found out the antisense FtsZ gene can be used to control the shape of E.coli's colony.
2012-4-9 Mon
Shen Gao: Choose Pet-32a plasmid for our experiment.
2012-4-12 Thur
Xiaodan Xu: Found a way to verification the primers.
2012-4-13 Fri
Xintong Ling: Finished the simple cell division model.
2012-4-17 Tue
Shen Gao: Learnt about the primer design.
Xingyu Zhou: Designed the quorum sensing part——the AHL system.
Muyuan Chen: Use vectorNTI to analysis plasmid.
2012-4-19 Thur
Xintong Ling:finished the Cellular Automata based on the AHL density.
Xingyu Zhou:Made the expriment plan for the AHL system and found some related kits for his plan.
2012-4-20 Fri
Muyuan Chen: Made the experiment plan for the antisense FtsZ.
Xintong Ling:improved the model of cellular automata.
2012-4-22 Sun
Shen Gao:improved the method of gel extraction and agarose gel electrophoresis.
Xingyu Zhou:Made a summary of AHL sensing system.
2012-4-24 Tue
Xintong Ling & Kai Gui:made the schedule for the further modeling.
Kai Gui:finished the model of biosensor.
2012-4-26 Thur
Xiaodan Xu:Made the experiment plan for antisense FtsZ.
2012-4-29 Sun
Xiaodan Xu:Made a list of the protection base for the restriction sites.
May
2012-5-2 Wed
Xintong Ling: modified the model of biosensor.
2012-5-3 Thur
Xiaodan Xu:Chose the target fragment of FtsZ.
2012-5-7 Mon
Muyuan Chen & Xiaodan Xu: Modified the experiment program--the promoter and the hairpin of the antisense fragment.
2012-5-8 Tue
Ying Cheng: Made the summary of the previous project.
Kai Gui: Finished the model of cellular automata based on the AHL signal diffusion.
2012-5-10 Thur
Our group discussed about the human practice.
Xingyu Zhou:Modified the experiment program of AHL part.
Xintong Ling: Summarise the previous model.
Xiaodan Xu: sorted out the kits list.
2012-5-11 Fri
Xintong Ling:modified the model of cellular automata.
2012-5-12 Sat
Xingyu Zhou:finished the complete AHL experiment program.
2012-5-13 Sun
Xiaodan Xu: Made a list of possibly useful kits.
2012-5-14 Mon
Muyuan Chen: Ordered the paired termini sequence.
2012-5-16 Wed
Xintong Ling & Kai Gui & Jun Zhang: had a meeting talking about the model.
2012-5-22 Tue
Xingyu Zhou: Found out the chemical way of the AHL detection.
2012-5-24 Thur
Xiaodan Xu: transformed the K082034 to make sure the method of Kit usage is correct.
2012-5-27 Sun
Had a group meeting and summarise the work we had done.Made the schedule for the summer vacation and everyone started the preparation for their final examinations.
2012-5-31 Thur Xingyu Zhou: Finished the budget for our project.
June Week 3
2012-6-12 Tue
We got the paired termini fragment in the PUC vector.
2012-6-16 Sat
Muyuan Chen:wrote the experiment program of AHL.(BBa_T9002+F1610)
June Week 4
2012-6-18 Mon
PCR: FtsZ target fragment
Transform: F1610 R0010
Recovery: I13521
2012-6-19 Tue
Check: F1610 failed; R0010 growed
Digestion: FtsZ1 FtsZ5 paired termini
Transform: F1610
Culture: R0010
Results: AHL failed
2012-6-20 Wed
Check: F1610 failed again
Transform FtsZ1/5+paired termini
Results: AHL failed
2012-6-21 Thur
Check: None
Miniprep plasmids R0010
Transform FtsZ1/5+paired termini
Culture: F1610; C0261; B0015
Others Found our instructor Zuhong Lu and reported our schedule.
2012-6-22 Fri
Check: F1610 FtsZ5+paired termini failed
PCR: Paired termini
Transform: J5526; I13202; T9002
Culture: FtsZ1+paired termini; T9002
Results: AHL failed; We got ftsZ1+termini~
2012-6-23 Sat
Check: J5526 I13202 growed.
Miniprep plasmids T9002
Culture: J5526; I13202
Others Muyuan Chen : Designed the toggle switch. BBa_K415301 BBa_K415300
June Week 5
2012-6-25 Mon
Miniprep plasmids: I13202; J5526
Transform: I13202; K415023; K091230
Recovery: Pet32a
Culture: I13202+T9002
2012-6-26 Tue
Check: K415023 failed
Digestion: T9002; I13202
Transform: K415023
Recovery: I13202
Culture: I13202; K091230
Results: Toggle switch failed
2012-6-27 Wed
Digestion: T9002+I13202
Transform: R0082; K415301; M30109; K415023; K091230
Recovery: T9002
2012-6-28 Thur
Check: T9002 failed
Ligation: T9002+I13202
Recovery: J5526
Culture: R0082 K415301 M30109 K415023 K091230
Results: AHL maybe failed.
2012-6-29 Fri
Check: R0082 K415023 failed
Miniprep plasmids: K415301; M30109; K091230
Digestion: Pet32a; paired termini
Ligation: Pet32a+paired termini
Transform: R0082; R0083; R0084
Results: AHL maybe would failed.
2012-6-30 Sat
Check: R0082 R0083 R0084 failed
Miniprep plasmids: Pet32a
Digestion: ftsZ +paired termini; Pet32a
Ligation: ftsZ+paired termini+pet32a
Recovery: Pet32a+egfp
Culture: FtsZ1 +Paired termini
Results: Light sensor failed

 "
"