Team:Peking/Modeling/Luminesensor
From 2012.igem.org
Summary
We have managed conduct modeling to guide the optimization of our Luminesensor, combining protein kinetics, thermodynamics, and stochastic simulation with molecular docking together. The modeling results point out two critical mutation sites (74 & 135) on VVD, the dimerization domain and a mutant choise of LexA, the DNA binding domain of our Luminesensor. The modeling results also shows the system still works well even if considering noise and endogenous competition.
Kinetic Network
Firstly we established a reaction network for the DNA binding process of Luminesensor. Previous works by other scientists indicate that the Vivid (VVD) protein, a sensing domain of Luminesensor, dimerizes in the presence of light,[1] and the LexA protein, a binding domain of Luminesensor, binds at specific sequences on DNA predominantly when coupled[2] and subsequently represses the interest gene. By combining with the mechanisms of the two functional domains of Luminesensor, we concluded with the following kinetic network:
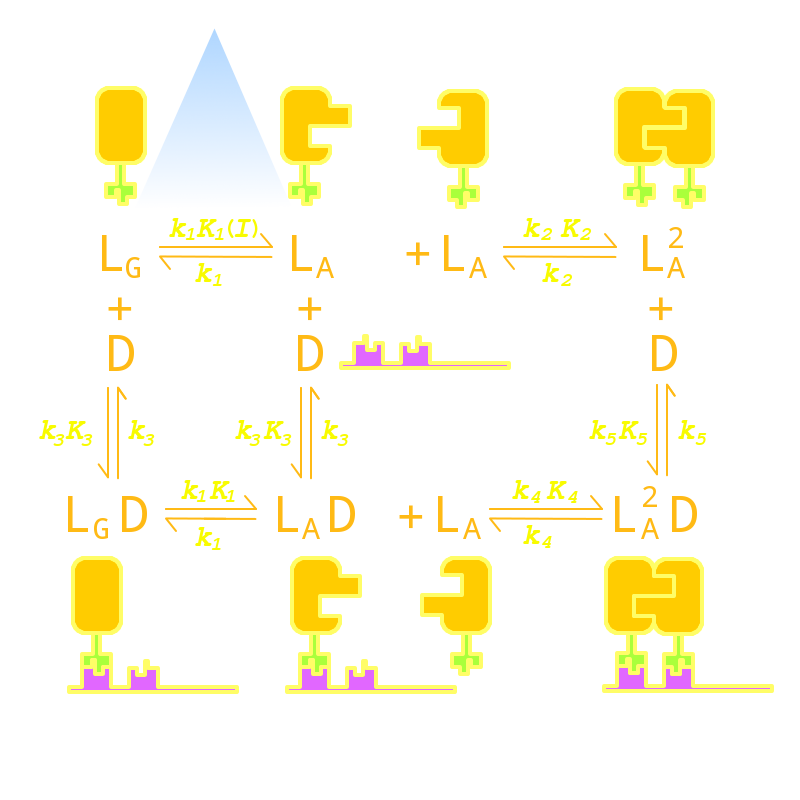
Fig 1. Kinetic Network of our Luminesensor
where
- L : Luminesensor,
- LG : the ground state --- with its VVD N-cap locked,
- LA : the active state --- with its VVD N-cap released,
- DL : the specific DNA binding site to Luminesensor.
Since no multi-intermediate reactions are hidden in the network above, all reactions can be regarded as elementary reactions.
Reference
- 1. Zoltowski, B.D., Crane, B.R.(2008)Light Activation of the LOV Protein Vivid Generates a Rapidly Exchanging Dimer.Biochemistry, 47: 7012: 7019
- 2. Mohana-Borges, R., Pacheco, A.B., Sousa, F.J., Foguel, D., Almeida, D.F., and Silva, J.L. (2000). LexA repressor forms stable dimers in solution. The role of specific DNA in tightening protein-protein interactions. J. Biol. Chem., 275: 4708: 4712
 "
"














