Team:NTNU Trondheim/Yeast
From 2012.igem.org
| Line 8: | Line 8: | ||
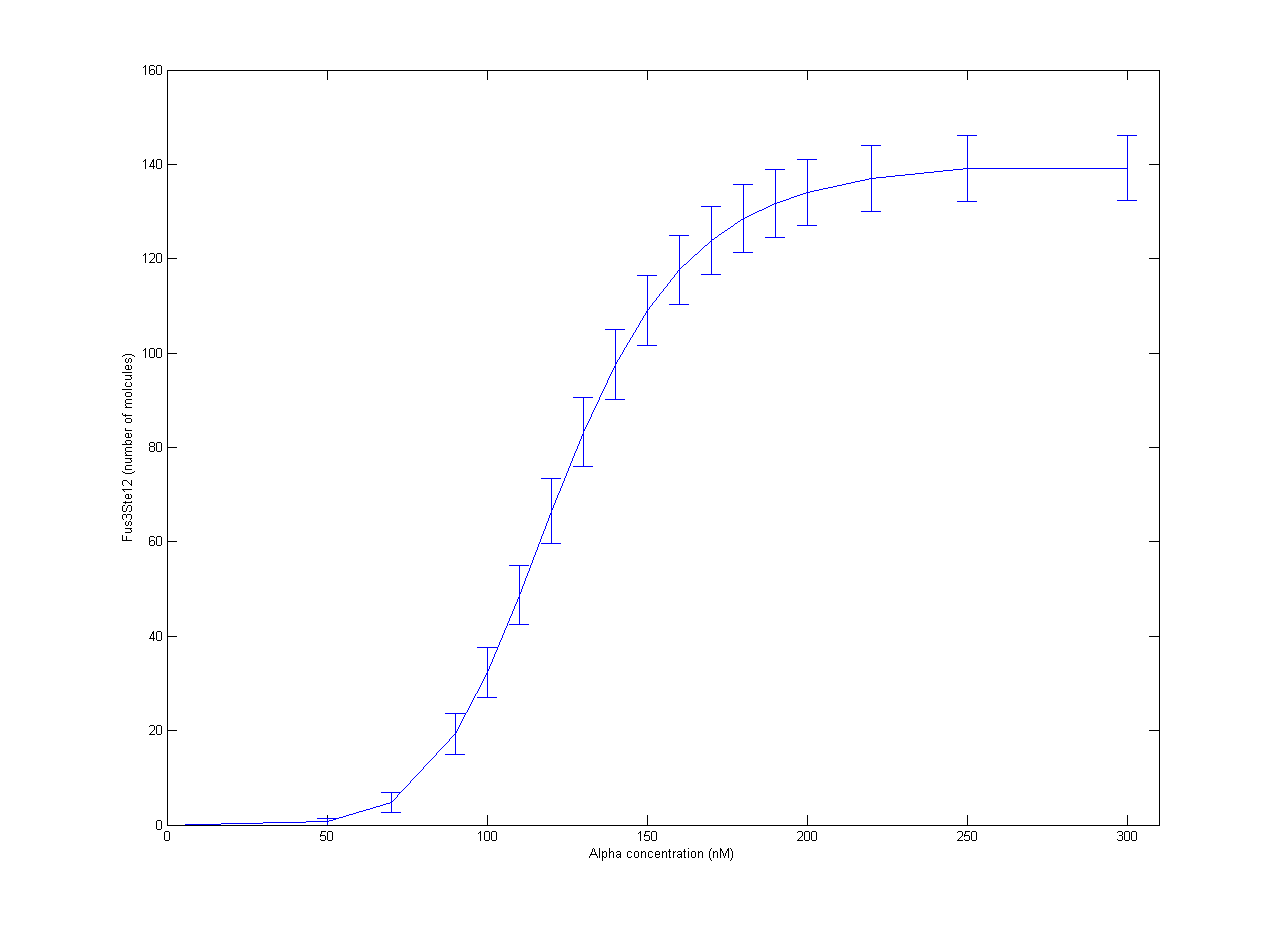
Our part of the collaboration with RHiT was helping them with stochastic modelling of the trigger system for mating in yeast. The full mechanism has been quite well studied, but it is very complicated[http://dx.doi.org/10.1002/yea.1122]. In RHiTs [[Team:RHIT/Modeling|model]], the final steps of the mechanism is activation of the Ste12 protein by the Fus3 enzyme. To simplify the model, production of Fus3 in the model was described by a sigmoid curve found in experiments[http://dx.doi.org/10.1038/nature08946] with respect to the concentration of alpha pheromone. Inactive Ste12 was quickly activated by the presence of Fus3, so the outcome of active Ste12 followed a similar sigmoid curve, giving the expected switch behaviour. The resulting plot is shown in Figure 1. Each point is the average of 100 trajectories with the error bars indicating one standard deviation. | Our part of the collaboration with RHiT was helping them with stochastic modelling of the trigger system for mating in yeast. The full mechanism has been quite well studied, but it is very complicated[http://dx.doi.org/10.1002/yea.1122]. In RHiTs [[Team:RHIT/Modeling|model]], the final steps of the mechanism is activation of the Ste12 protein by the Fus3 enzyme. To simplify the model, production of Fus3 in the model was described by a sigmoid curve found in experiments[http://dx.doi.org/10.1038/nature08946] with respect to the concentration of alpha pheromone. Inactive Ste12 was quickly activated by the presence of Fus3, so the outcome of active Ste12 followed a similar sigmoid curve, giving the expected switch behaviour. The resulting plot is shown in Figure 1. Each point is the average of 100 trajectories with the error bars indicating one standard deviation. | ||
| - | The equations used in the model are given in the table below. | + | The equations used in the model are given in the table below. The parameters are taken from[http://dx.doi.org/10.1002/yea.1122 [1<nowiki>]</nowiki>] and are modelled using mass action solvers, except Fus3 → Fus3PP, which use a sigmoid function. |
| + | |||
{|border="1" | {|border="1" | ||
| Line 15: | Line 16: | ||
!Propensity | !Propensity | ||
!Comment | !Comment | ||
| + | |- | ||
| + | |Fus3 → Fus3PP | ||
| + | |* | ||
| + | |Activation of Fus3 | ||
| + | |- | ||
| + | |Fus3PP → Fus3 | ||
| + | |150 | ||
|} | |} | ||
Revision as of 11:59, 14 August 2012
 "
"