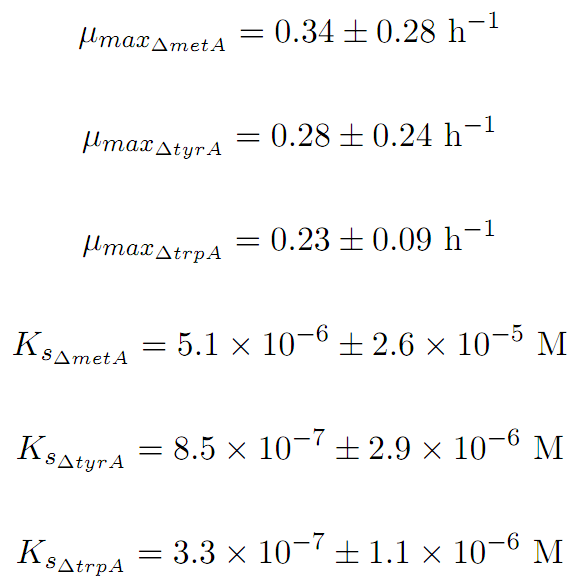Team:British Columbia/Consortia
From 2012.igem.org
m (→Description:) |
|||
| Line 6: | Line 6: | ||
| - | The three interdependent ecoli auxotrophs studied in our project are: | + | The three interdependent ecoli auxotrophs studied in our project are: MetA-, TrpA-, and TyrA- knock outs. This model is designed to predict the consortium’s population dynamics, based on the measured amino acid excretion rates without tuning them. However, the code of the model is written in a way that when we decide to tune the population by adding in different concentrations of inducers, we can simply update the excretion and consumption rate equations. Thus this model is valid for both situations. It can even be generalized to be used by any interdependent ecoli consortium when Met, Trp, and Tyr are the limiting nutrients/substrates for their growth, with some modifications. |
| + | |||
| + | The key assumption for this model is that the Methionine, Tryptophan, and Tyrosine are the only growth limiting substrates for MetA-, TrpA-, and TryA- ecoli knock outs respectively. | ||
| + | |||
| + | |||
| + | |||
Revision as of 00:01, 29 September 2012

Description:
The three interdependent ecoli auxotrophs studied in our project are: MetA-, TrpA-, and TyrA- knock outs. This model is designed to predict the consortium’s population dynamics, based on the measured amino acid excretion rates without tuning them. However, the code of the model is written in a way that when we decide to tune the population by adding in different concentrations of inducers, we can simply update the excretion and consumption rate equations. Thus this model is valid for both situations. It can even be generalized to be used by any interdependent ecoli consortium when Met, Trp, and Tyr are the limiting nutrients/substrates for their growth, with some modifications.
The key assumption for this model is that the Methionine, Tryptophan, and Tyrosine are the only growth limiting substrates for MetA-, TrpA-, and TryA- ecoli knock outs respectively.
Constants:
Some of the key constants for the Monod Kinetics are obtained through analyzing various growth rates under different known amino acid concentrations. The values are listed as follows:
Equations:
 "
"