Team:Amsterdam/project/growthrates
From 2012.igem.org
(Difference between revisions)
(→Growth curve experiments) |
|||
| Line 17: | Line 17: | ||
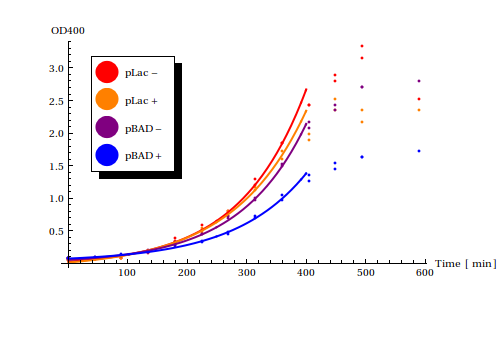
[[File:rateestimation.png|thumb|Growth rates of two different constructs (pLac, pBAD) with either the corresponding signal (lactose, arabninose) present or not present]] | [[File:rateestimation.png|thumb|Growth rates of two different constructs (pLac, pBAD) with either the corresponding signal (lactose, arabninose) present or not present]] | ||
| - | |||
Using the Mathematica function <code>NonLinearModelFit</code> functions of the form <math>a + b e^{c t}</math>, with <math>t</math> as time in minutes, were fitted to the exponential phases of the growth curves. | Using the Mathematica function <code>NonLinearModelFit</code> functions of the form <math>a + b e^{c t}</math>, with <math>t</math> as time in minutes, were fitted to the exponential phases of the growth curves. | ||
| - | <table | + | <table align="center"><tr style="background-color:orange;"><td>Strain</td><td>Function</td></tr> |
<tr><td>pLac + IPTG</td><td><math>-0.0501829+0.0743786 e^{0.0089986 t}</math></td></tr> | <tr><td>pLac + IPTG</td><td><math>-0.0501829+0.0743786 e^{0.0089986 t}</math></td></tr> | ||
<tr><td>pLac - IPTG</td><td><math>-0.0823022+0.0938526 e^{0.00812998 t} </math></td></tr> | <tr><td>pLac - IPTG</td><td><math>-0.0823022+0.0938526 e^{0.00812998 t} </math></td></tr> | ||
| Line 28: | Line 27: | ||
</table> | </table> | ||
| + | Although no control strain was grown to compare the effects of the constructs to the wild-type strains, we can assume that all constructs slow down the growth rates somewhat. | ||
| + | For both pLac and pBAD, induction of the plasmid with the corresponding signal result in slower growth rates compared to uninduced growth rates. | ||
| + | Suprisingly, pBAD reduces the growth rate more strongly than pLac. | ||
| + | pLac is known to be a better stronger promoter than pBAD with higher leaky expression rates and was thus expected to have a stronger negative effect on the growth rate. | ||
| + | This results suits us very well! As you can read <here> pBAD was also shown to have lower leaky expression rates in our own experiments and thus functions more robustly. | ||
</div> | </div> | ||
</div> | </div> | ||
{{Team:Amsterdam/Foot}} | {{Team:Amsterdam/Foot}} | ||
Revision as of 09:43, 23 September 2012
 "
"