Team:Amsterdam/project/growthrates
From 2012.igem.org




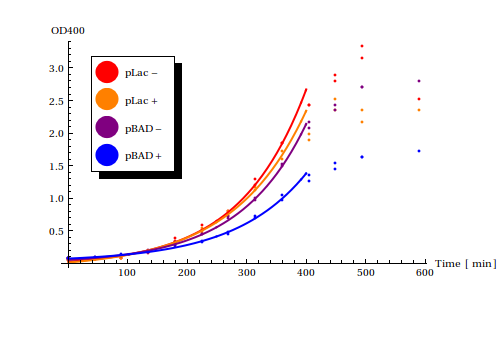
Calculated growth rates from growth curve experiments
The division rate of the cells is of vital importance to the duration in which the registered signal is stored in the cell. The slower the growth rate of the Cellular Logbook, the slower the dilution of the methylated plasmids and hence the longer the stored signal is retained. The impact on the growth rates of the various constructs we have gives an indication of their transcriptional burden.
This is why We´ve performed growth experiments for two constructs: pSB1AT3-pLac-MTase and pSB1AT3-pBAD-MTase. At this time we didn't have our more complete construct including the Zinc Finger prepared yet. Both experiments were performed in cell strain <math>\text{DH5}\alpha</math>.
Using the Mathematica function NonLinearModelFit functions of the form <math>a + b e^{c t}</math>, with <math>t</math> as time in minutes, were fitted to the exponential phases of the growth curves.
| Strain | Function | Average replication time |
|---|---|---|
| pLac + IPTG | <math>-0.0501829+0.0743786 e^{0.0089986 t}</math> | <math>\frac{ln(2)}{0.0089986} = 77\ \text{mins}</math> |
| pLac - IPTG | <math>-0.0823022+0.0938526 e^{0.00812998 t} </math> | <math>\frac{ln(2)}{0.00812998} = 85\ \text{mins}</math> |
| pBAD + Arabinose | <math>-0.0101962+0.0614008 e^{0.00889096 t} </math> | <math>\frac{ln(2)}{0.0089096} = 78\ \text{mins}</math> |
| pBAD - Arabinose | <math>-0.0233633+0.0489266 e^{0.00830895 t} </math> | <math>\frac{ln(2)}{0.00830896} = 83\ \text{mins}</math> |
Discussion
Although no control strain was grown to compare the effects of the constructs to the wild-type strains, we can assume that all constructs slow down the growth rates somewhat. For both pLac and pBAD, induction of the plasmid with the corresponding signal result in slower growth rates compared to uninduced growth rates. Suprisingly, pBAD reduces the growth rate more strongly than pLac. pLac is known to be a better stronger promoter than pBAD with higher leaky expression rates and was thus expected to have a stronger negative effect on the growth rate.
This results suits us very well! As you can read <here> pBAD was also shown to have lower leaky expression rates in our own experiments and thus functions more robustly.
 "
"