Team:Shenzhen/Result/YAO.Channel
From 2012.igem.org
-
S
BioBricks
- Summary
- YAO.Genome
- YAO.Channel
- YAO.Sensor
- YAO.Suicider
-
p
Notebook
- Team History
- YAO.Genome
- YAO.Channel
- YAO.Sensor
- YAO.Suicider
- YAO.Factory
-
e
Practices



- 1. SP of Zim17
- 2. SP of ferredoxi
- 3. SP of Tim21
- 4. SP of Tom40
- 5. SP of Tom70
- 6. SP of Tom22
- Causes
- Solution
- Protocol
- Purpose
- Materials
- Methods
- I. First Vector Construction
- II. Second Vector Construction
- III. Third Vector Construction
- IV. Construction of yeast expression vector
- V. Yeast transformation
Construction of a Signal Peptide System
Signal peptides are the tools we use to assemble YAO. Channel-Toc and Tic proteins, and to guide the protein into YAO matrix through YAO. Channel. We have designed several Signal Peptide BioBricks (shown in http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2012&group=Shenzhen).
In total, there are 6 types of signal peptide involved in our project: signal peptide (SP) of Zim17, SP of ferredoxi, SP of Tim21, SP of Tom40, SP of Tom70, SP of Tom22.
Zim17 is a zinc finger protein located in mitochondrial matrix. The signal peptide of Zim17 is amino-terminal segments rich in positively charged residues that form two to three turns of a helix with amphipathic character, and it directs the proteins imported into mitochondrial matrix. The sequence can be found at http://www.uniprot.org/blast/?about=P42844[1-47]. We synthesized the sequence and linked it with RFP & YFP.
The document we mainly refer to is:
Burri L, Vascotto K, Fredersdorf S, Tiedt R, Hall MN, Lithgow T. Zim17, a novel zinc finger protein essential for protein import into mitochondria. J. Biol. Chem. 2004;279:50243-50249.
The signal peptide of ferredoxi, as a chloroplast-target transit peptide, has an α helix at its N-terminus, followed by an unstructured C-terminal domain of ~19 amino acids. The sequence can be found at http://www.uniprot.org/blast/?about=P07839%5b1-32%5d. We synthesized the sequence and linked it with RFP & YFP.
The document we mainly refer to is:
Bruce, B. D. (2000). "Chloroplast transit peptides: structure, function and evolution." Trends in Cell Biology 10(10): 440-447.
Tim21 is one component of the translocase of the inner mitochondrial membrane (TIM complex), and the signal peptide of Tim21 contains 239 amino acids with a N-terminally single transmembrane segment showing the characteristics of a positively charged mitochondrial presequence with a predicted cleavage site after residue 42. The sequence can be found at http://www.uniprot.org/blast/?about=P53220[1-70]. We synthesized the sequence and linked it with RFP & YFP.
The document we mainly refer to is:
Chacinska, A., M. Lind, et al. (2005). "Mitochondrial Presequence Translocase: Switching between TOM Tethering and Motor Recruitment Involves Tim21 and Tim17." Cell 120(6): 817-829.
Tom40 is the core component of the translocase of the outer mitochondrial membrane (TOM complex), presenting an β-barrel structure. The signal peptide of Tom40 is a conserved β-signal motif, located near the C-terminus, The sequence can be found in the document BMC Genomics, 12, 79. We synthesized the sequence and linked it with RFP & YFP.
The document we mainly refer to is:
Imai, K., Fujita, N., Gromiha, M.M. and Horton, P. (2011) Eukaryote-wide sequence analysis of mitochondrial beta-barrel outer membrane proteins. BMC Genomics, 12, 79.
Tom70 is also one component of TOM complex, but only contain a single transmembrane segment at their N terminus. The signal peptide of Tom70 is the transmembrane domain comprising a hydrophilic, positively charged segment. The sequence can be found in http://www.uniprot.org/blast/?about=P07213[11-30]. We synthesized the sequence and linked it with RFP & YFP.
The document we mainly refer to is:
Rapaport, D. (2003). "Finding the right organelle." EMBO Rep 4(10): 948-952.
Tom22 is similar with Tom70, but it’s a tail-anchored protein. The signal peptide of Tom22 is also similar with the one of Tom70: moderately hydrophobic, relatively short, with positive charges at the flanking regions. The sequence can be found in http://www.uniprot.org/blast/?about=P49334[98-119]. We synthesized the sequence and linked it with RFP & YFP.
The document we mainly refer to is:
Plant Physiology July 2000 vol. 123 no. 3 811-816.
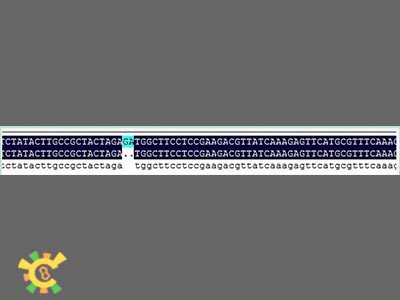
Construction of Signal Peptide System
Due to some inaccurate information in the iGEM website and our own mistakes, some errors accrued when we attempted to link the reporter gene with signal peptide.
The followings are the reason why the errors took place and the approach we applied to solved the problem.
The general adapter is:
GAATTC GCGGCCGC T TCTAGA G[ATG ... TAA TAA] T ACTAGT A GCGGCCG CTGCAG
And general adapter of fusion protein is:
GAATTC GCGGCCGC T TCTAGA [ATG ... TAA TAA] ACTAGT A GCGGCCG CTGCAG
But our adapter of signal peptide is:
GAATTC GCGGCCGC T TCTAG [ATG ... TAA TAA] T ACTAGT A GCGGCCG CTGCAG
Bad news came when we attempted to link the General adapter with the General adapter fragment. The fusion protein is as following:
GAATTC GCGGCCGC T TCTAGA G[part1] T ACTAGA G [part2]T ACTAGT A GCGGCCG CTGCAG
As we can see, there are 8 bases at the fused locus, and in this case, frame shift mutation will occur.
We also produce fusion protein using two General adapter, the result is:
GAATTC GCGGCCGC T TCTAGA [part1] ACTAGA [part2] ACTAGT A GCGGCCG CTGCAG
There are 6 bases at the fused locus. No stop codon occurs, and it will not cause frame shift mutation.
If we use our adapter to link the adapter we designed, the locus is:
GAATTC GCGGCCGC T TCTAG [part1] CTACTAGAG [part2] T ACTAGT A GCGGCCG CTGCAG
In this case, no frame shift mutation occurs, and there are a lot of GFPs available if we select this adapter for use.
However, when sequencing the fusion protein, we found the fusion locus had lost 2 bases thus causing frameshift mutation, and that the adapter of E1010 linked by backbone PSB1A3 became:
GAATTC GCGGCCGC T TCTAG [ATG ... TAA TAA] T ACTAGT A GCGGCCG CTGCAG
To correct this error, we designed some primers to perform the point mutation.
Table 1. Primers we used in this experiment
Common | 5'-GTTTCTTCGAATTCGCGGCCGCTTCTAG |
ZIM17-R | 5'-GTTTCTTCCTGCAGCGGCCGCTATACTAGTAGATGTGCGATGAT |
FER-R | 5'-GTTTCTTCCTGCAGCGGCCGCTATACTAGTAGGGCCATGCAGCT |
TIM21-R | 5'-GTTTCTTCCTGCAGCGGCCGCTATACTAGTAGGGATTTTACTTG |
TOM22-R | 5'-GTTTCTTCCTGCAGCGGCCGCTATACTAGTAGCGGCAAGTATAG |
TOM70-R | 5'-GTTTCTTCCTGCAGCGGCCGCTATACTAGTAGGTTGTAATAATA |
TOM20-R | 5'-GTTTCTTCCTGCAGCGGCCGCTATACTAGTAGCCTGAATTGCGG |
TOM40-R | 5'-GTTTCTTCCTGCAGCGGCCGCTATACTAGTAGCAATTGAGGAAG |
COM-F is the forward primer of all signal peptides, and the other seven are the reverse primers of signal peptides. After our experimental adapters are as followings:
GAATTC GCGGCCGC T TCTAG [part] CT ACTAG TA T A GCGGCCG CTGCAG
We add three bases in the adapter. If we link the signal peptide at N-terminal, the fused locus is:
GAATTC GCGGCCGC T TCTAG [part1] CTACTAGAG [part2]T ACTAGT A GCGGCCG CTGCAG
In this case, there are nine bases at the locus, and there aren’t stop codon or frame shift mutations, and our adapter can be used.
After such treatment, final adapter is:
GAATTC GCGGCCGC T TCTAGA G [part] CT ACTAG TA T A GCGGCCG CTGCAG
If we add signal peptide at the N-terminal of adapter, fusion protein is:
GAATTC GCGGCCGC T TCTAGAG [part1] CTACTAGAG [part2] CT ACTAGT ATA GCGGCCG CTGCAG
Step 1:Extraction plasmid
Zim17, FER, TIM21, TOM22, TOM70, TOM20, TOM50
Step 2:Point mutation
PCR system(take ZIM17 as example):
ZIM17 plasmid template 0.2ul
Ex Taq 0.5ul
10 x Ex Taq Buffer 2.0ul
dNTP Mix 1.0ul
COM-Fv0.5ul
ZIM17-R 0.5ul
ddH2O 15.3ul
Reaction condition:
94℃ 5min
94℃v30s
55℃ 30s 30 cycle
72℃ 30s
72℃ 10min
12℃ ∞
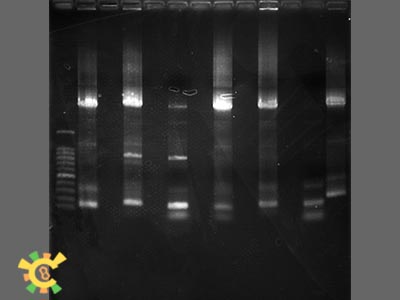
Step 3:Add 4ul, 6 x loading Buffer, and Run agarose gel. The result are as followings:
Step 4:gel extraction(use QIAquick Gel Extraction Kit)
Step 5:use NANODROP 2000 to measure concentration.
Step 6: enzyme digestion
enzyme digestion system:
target fragment 8ul
ECoRI 0.5ul
SpeI 0.5u
10 x H Buffer 1ul
Reaction condition: 37℃ x 3h + 70℃ x 30min
Signal Peptides Validation Protocol
Verify that the mitochondrial signal peptides can lead peptides into the mitochondria, or onto the mitochondrial membrane, while the chloroplast signal peptides can not.
GFP, Signal peptides (ZIM17, FER, TIM21, TOM22, TOM70, TOM20, TOM40), yeast strain.
1. Synthesis of ZIM17, FER, TIM21, TOM22, TOM70, TOM20, TOM40.
2. The synthetic fragments are connected with PUC57 the carrier. Mix the strain which containing the carrier and 4 ml of LB+AMP liquid medium in 15 ml centrifuge tube, shake at 37 ℃, 220 rpm overnight.
3. Mix other two strains( BBa-E0030, BBa-E1010 containing biobrick) and 4 ml of LB+AMP liquid medium in 15 ml centrifuge tube, shake at 37 ℃, 220 rpm overnight.
4. Extract plasmid ( ZIM17, FER, TIM21, TOM22, TOM70, TOM20, TOM40, BBa-E0030, BBa-E1010).
5. Use NANODROP 2000 from Thermo Co. to measure plasmid concentration.
6. Digest:
ZIM17, FER, TIM21, TOM22, TOM70, TOM20, TOM40 2-3ug respectively.
ECoRI 2ul
SpeI 2ul
10 x H Buffer 5ul
ddH2O 50ul
BBa-E0030, BBa-E1010 2-3ug respectively
ECoRI 2ul
XbaI 2ul
10 x M Buffer 5ul
ddH2O 50ul
- Reaction conditions:37℃ x 3h + 70℃ x 30min
7. Run a 1% agarose gel 120V 45 min, and EB staining.
8. Gel Extraction( QIAquick Gel Extraction Kit).
9. Measure the concentration of the recycle fragments.
10. Ligation:
(1) ZIM17(ECoRI/SpeI) 6ul (2) ZIM17(ECoRI/SpeI) 6ul
BBa-E0030(ECoRI/XbaI) 2ul BBa-E1010(ECoRI/XbaI) 2ul
T4 ligase 1ul T4 ligase 1ul
T4 ligase Buffer 1ul T4 ligase Buffer 1ul
and the same to (3) ~ (14).
- Ligation condition: Room temperature, 3h.
11. Transformation
(1) UV sterilization half an hour earlier, and melt competent cells (DH5a) with ice bath.
(2) 10 μl ligation product + 50 μl of competent cells, with 30 min ice bath.
(3) 42 ℃ water bath heat shock 50s.
(4) cool down with ice for 2 min.
(5) Adding 500μl LB liquid medium without antibiotics, 37 ℃ shake (225 rpm) 1 h, recovery.
(6) 3000 rpm × 5 min, discard 400μl supernatant and percussion of the remaining liquid and mix.
(7) Overlay 60μl broth on solid LB medium with Kan resistance, 37 ℃, inverted overnight culture in incubator.
12. Validation positive clones
dNTP mix (2.5mM) 1μl
PCR 10×buffer 2μl
VF 0.5μl
VR 0.5μl
Ex Taq 0.5μl
H2O 15.5μl
Primer sequence:
VF: TGCCACCTGACGTCTAAGAA
VR: ATTACCGCCTTTGAGTGAGC
94℃ 3 min
94℃ 30s
55℃ 30s 30 cycles
72℃ 1 min
72℃ 10 min
12℃ ∞
1% to 1.5% agarose gel electrophoresis, voltage <130, time ≈ 40 min.
- Result:
13. Sequencing
Pick single colony of PCR positive clones to sequencing
14. Named the correct vectors( for convenient description)
ZIM17-E0030, ZIM17-E1010, FER-E0030, FER-E1010, TIM2-E0030, TIM2-E1010, TOM22-E0030, TOM22-E1010, TOM70-E0030, TOM70-E1010, TOM20-E0030, TOM20-E1010, Tom40-E0030, Tom40-E1010
1. Mix the strain which containing following carrier and 4 ml of LB+AMP liquid medium in 15 ml centrifuge tube, shake at 37 ℃, 220 rpm overnight.
ZIM17-E0030, ZIM17-E1010, FER-E0030, FER-E1010, TIM2-E0030, TIM2-E1010, TOM22-E0030, TOM22-E1010, TOM70-E0030, TOM70-E1010, TOM20-E0030, TOM20-E1010, Tom40-E0030, Tom40-E1010(kan resistant), T-gz-GAL1(Amp resistant).
2. Extract plasmid
3. Measure plasmid concentration
4. Digestion
ECoRI and XbaI:
ZIM17-E0030, ZIM17-E1010, FER-E0030, FER-E1010, TIM2-E0030, TIM2-E1010, TOM22-E0030, TOM22-E1010, TOM70-E0030, TOM70-E1010, TOM20-E0030, TOM20-E1010, Tom40-E0030, Tom40-E1010
ECoRI and SpeI:
T-gz-GAL1
5. Gel electrophoresis, recycle and measure plasmid concentration
6. Ligation
ZIM17-E0030, ZIM17-E1010, FER-E0030, FER-E1010, TIM2-E0030, TIM2-E1010, TOM22-E0030, TOM22-E1010, TOM70-E0030, TOM70-E1010, TOM20-E0030, TOM20-E1010, Tom40-E0030, Tom40-E1010 2ul
respectively ligating with T-gz-GAL1 6ul.
7. Transformation
8. PCR
9. Shake culture
10. Sequencing
11. Named the correct vectors( for convenient description)
GAL-ZIM17-E0030, GAL-ZIM17-E1010, GAL-FER-E0030, GAL-FER-E1010, GAL-TIM2-E0030, GAL-TIM2-E1010, GAL-TOM22-E0030, GAL-TOM22-E1010, GAL-TOM70-E0030, GAL-TOM70-E1010, GAL-TOM20-E0030, GALTOM20-E1010, GAL-Tom40-E0030, GAL-Tom40-E1010.
1. Mix the strain which containing following carrier and 4 ml of LB+AMP liquid medium in 15 ml centrifuge tube, shake at 37 ℃, 220 rpm overnight.
GAL-ZIM17-E0030, GAL-ZIM17-E1010, GAL-FER-E0030, GAL-FER-E1010, GAL-TIM2-E0030, GAL-TIM2-E1010, GAL-TOM22-E0030, GAL-TOM22-E1010, GAL-TOM70-E0030, GAL-TOM70-E1010, GAL-TOM20-E0030, GAL-TOM20-E1010, GAL-Tom40-E0030, GAL-Tom40-E1010(kan resistant)
BBa-J63002(Amp resistant, yeast ADH terminator)
2. Extract plasmid
3. Measure plasmid concentration
4. Digestion SpeI and PstI with 10X H Buffer:
GAL-ZIM17-E0030, GAL-ZIM17-E1010, GAL-FER-E0030, GAL-FER-E1010, GAL-TIM2-E0030, GAL-TIM2-E1010, GAL-TOM22-E0030, GAL-TOM22-E1010, GAL-TOM70-E0030, GAL-TOM70-E1010, GAL-TOM20-E0030, GAL-TOM20-E1010, GAL-Tom40-E0030, GAL-Tom40-E1010
XbaI and PstI with 10X H Buffer:
BBa-J63002
5. Gel electrophoresis, recycle and measure plasmid concentration
6. Ligation
GAL-ZIM17-E0030, GAL-ZIM17-E1010, GAL-FER-E0030, GAL-FER-E1010, GAL-TIM2-E0030, GAL-TIM2-E1010, GAL-TOM22-E0030, GAL-TOM22-E1010, GAL-TOM70-E0030, GAL-TOM70-E1010, GAL-TOM20-E0030, GAL-TOM20-E1010, GAL-Tom40-E0030, GAL-Tom40-E1010 2ul respectively ligating with BBa-J63002 6ul.
7. Transformation
8. PCR
9. Shake culture
10. Sequencing
11. Named the correct vectors( for convenient description)
GAL-ZIM17-E0030-J63002, GAL-ZIM17-E1010-J63002, GAL-FER-E0030-J63002, GAL-FER-E1010-J63002, GAL-TIM2-E0030-J63002, GAL-TIM2-E1010-J63002, GAL-TOM22-E0030-J63002, GAL-TOM22-E1010-J63002, GALTOM70-E0030-J63002, GAL-TOM70-E1010-J63002, GAL-TOM20-E0030-J63002, GALTOM20-E1010-J63002, GAL-Tom40-E0030-J63002, GAL-Tom40-E1010-J63002
1. Mix the strain which containing following carrier and 4 ml of LB+AMP liquid medium in 15 ml centrifuge tube, shake at 37 ℃, 220 rpm overnight
GAL-ZIM17-E0030-J63002, GAL-ZIM17-E1010-J63002, GAL-FER-E0030-J63002, GAL-FER-E1010-J63002, GAL-TIM2-E0030-J63002, GAL-TIM2-E1010-J63002, GAL-TOM22-E0030-J63002, GAL-TOM22-E1010-J63002, GALTOM70-E0030-J63002, GAL-TOM70-E1010-J63002, GAL-TOM20-E0030-J63002, GALTOM20-E1010-J63002, GAL-Tom40-E0030-J63002, GAL-Tom40-E1010-J63002(kan resistant)
YEP352(Amp resistant, yeast ADH terminator)
2. Extract plasmid
3. Measure plasmid concentration
4. Digestion
XbaI and PstI:
GAL-ZIM17-E0030-J63002, GAL-ZIM17-E1010-J63002, GAL-FER-E0030-J63002, GAL-FER-E1010-J63002, GAL-TIM2-E0030-J63002, GAL-TIM2-E1010-J63002, GAL-TOM22-E0030-J63002, GAL-TOM22-E1010-J63002, GALTOM70-E0030-J63002, GAL-TOM70-E1010-J63002, GAL-TOM20-E0030-J63002, GAL-TOM20-E1010-J63002, GAL-Tom40-E0030-J63002, GAL-Tom40-E1010-J63002, YEP352
5. Gel electrophoresis, recycle and measure plasmid concentration
6. Ligation
GAL-ZIM17-E0030-J63002, GAL-ZIM17-E1010-J63002, GAL-FER-E0030-J63002, GAL-FER-E1010-J63002, GAL-TIM2-E0030-J63002, GAL-TIM2-E1010-J63002, GAL-TOM22-E0030-J63002, GAL-TOM22-E1010-J63002, GAL-TOM70-E0030-J63002, GAL-TOM70-E1010-J63002, GAL-TOM20-E0030-J63002, GAL-TOM20-E1010-J63002, GAL-Tom40-E0030-J63002, GAL-Tom40-E1010-J63002 6ul respectively ligating with YEP532 2ul.
7. Transformation
8. PCR
9. Shake culture
10. Sequencing
11. Named the correct vectors( for convenient description)
YEP-GAL-ZIM17-E0030-J63002, YEP-GAL-ZIM17-E1010-J63002, YEPGAL-FER-E0030-J63002, YEP-GAL-FER-E1010-J63002, YEP-GALTIM2-E0030-J63002, YEP-GAL-TIM2-E1010-J63002, YEP-GAL-TOM22-E0030-J63002, YEP-GAL-TOM22-E1010-J63002, YEP-GAL-TOM70-E0030-J63002, YEP-GAL-TOM70-E1010-J63002, YEP-GAL-TOM20-E0030-J63002, YEP-GAL-TOM20-E1010-J63002, YEPGAL-Tom40-E0030-J63002, YEP-GAL-Tom40-E1010-J63002
1. Extract plasmid
YEP-GAL-ZIM17-E0030-J63002, YEP-GAL-ZIM17-E1010-J63002, YEPGAL-FER-E0030-J63002, YEP-GAL-FER-E1010-J63002, YEP-GALTIM2-E0030-J63002, YEP-GAL-TIM2-1010-J63002, YEP-GAL-TOM22-E0030-J63002,
YEP-GAL-TOM22-E1010-J63002, YEP-GAL-TOM70-E0030-J63002, YEP-GAL-TOM70-E1010-J63002, YEP-GAL-TOM20-E0030-J63002, YEP-GAL-TOM20-E1010-J63002, YEPGAL-Tom40-E0030-J63002, YEP-GAL-Tom40-E1010-J63002
2. Preparation of yeast competent
(1) Streak a YPDA agar plate with a small portion of AH109 or Y187. Incubate the plate upside down at 30°C until colonies appear(~3days). Yeast strains can be stored for up to 1 month at 4°C on YPDA medium in culture plates.
(2) Prepare 1.1X TE/LiAc Solution.
(3) Prepare YPDA liquid medium (Yeast Protocols Handbook).
(4) Inoculate one colony(<4 weeks old, 2–3mm in diameter) into 3ml of YPDA medium in a sterile, 15-ml centrifuge tube.
(5) Incubate at 30°C with shaking for 8hr.
(6) Transfer 5µl of the culture to a 250-ml flask containing 50ml of YPDA.
(7) Incubate at 30°C with shaking at 230–250 rpm for 16–20hr.The OD600 should reach 0.15–0.3.
(8) Centrifuge the cells at 700 x g for 5min at room temperature.
(9) Discard the supernatant and resuspend the cell pellet in 100 ml of YPDA.
(10) Incubate at 30°C for 3–5hr(OD600=0.4~0.5).
(11) Centrifuge the cells at 700 x g for 5min at room temperature.
(12) Discard the supernatant and resuspend the cell pellet in 60ml of sterile, deionized H2O.
(13) Centrifuge the cells at 700 x g for 5min at room temperature.
(14) Discard the supernatant and resuspend the cells in 3 ml of 1.1 X TE/LiAc Solution.
(15) Split the resuspension between two 1.5-ml microcentrifuge tubes (1.5 ml pertube).
(16) Centrifuge each tube at high speed for 15 sec.
(17) Discard the supernatant and resuspend each pellet in 600 µl of 1.1 X TE/LiAc Solution.
3. Transformation
(1) 1.5ml centrifuge:
plasmid 0.2ug
Sperm DNA*(10 mg/ml) 0.1mg
Note: Sperm DNA: When freshly prepared water bath boil for 20 minutes, and immediately inserted into the ice bath and stored at -20 °C
(2) Add 100 ul with 1 × TE / LiAc resuspended yeast competent cells per tube, mixed with shaking;
(3) Each added 600ul PEG / LiAc, shaken vigorously (to improve the conversion efficiency), 30 °C, 200 rpm shaking culture 30 min;
(4) Each adding 70 ul DMSO, slowly inverted mix (not oscillating), 42 °C water bath thermal shock 15 min, insert into the ice bath to cool 1-2 min;
(5) 14,000rpm × 5 sec, discard supernatant, resuspend cells with 0.1 ml 1×TE, overlay on SD/-Ura solid medium, 30°C inverted culture 3 days in incubator.
4. Check out
Draw the colonies grown on the said plate in the galactose-containing SD/-Ura solid plates, until the colonies grow, single colonies were picked, and microscopic examination under a fluorescence microscope and record the results.
 "
"