Team:Austin Texas/ZombiE coli
From 2012.igem.org

Contents |
Human Practices
Modern science has enabled humanity to better combat many of the diseases that have plagued its history. Preventive care, such as vaccination, plays an especially important role in global healthcare. However, vaccination cannot provide a defense against presently unknown diseases, the "black swans" of the pathogenic world. Instead, education must be humanity's vanguard against an unprecedented outbreak of contagious disease.
Among the most common tropes in pop culture is the zombie outbreak. These apocalyptic scenarios often feature highly virulent pathogens transmitted via fluid contact. Recognizing the medium of transmission and avoiding it is definitely the most important step for survival in most zombie movies, and this message is highly applicable to real world disease prevention. Educating people on the nature of disease spread can greatly bolster their odds of survival if and when any such doomsday pathogen were to emerge.
Our ZombiE coli circuit represents an individual who, when exposed to an infectious agent, is transformed into a contagious host and disseminates further infectious agents into the surroundings. Upon infection, the individual takes upon a noticeable different appearance; in our model, this is change in color from fluorescent green to fluorescent red. We predict that the easily visualized color change spreading through a population of ZombiE coli will positively impact retention and further our take home message.
Our future goals on this project are to simulate the effects of immunity on the transmission of the infectious agents. Various types of resistance can be modeled, such as contagious carriers who exhibit no symptoms and innately immune individuals.
Project: ZombiE.coli
Introduction: Distinguishing Between E.coli populations
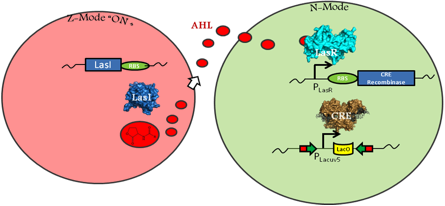
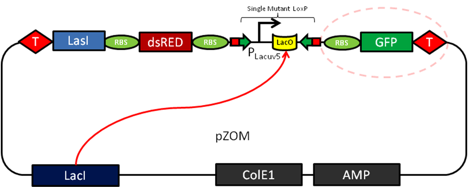
A circuit modeling transmissible disease based on a quorum sensing induced genetic switch must include certain E.coli behaviors. One allows for an infected, or zombie cell to infect normal cells, and one for a normal cell to irreversibly switch into a zombie cell. In order to easily distinguish between phenotypes of normal and zombie cells, different fluorescent proteins will be expressed in each cell population. A green fluorescent protein (GFP) will be expressed in normal cells, and a red fluorescent protein (RFP) expressed in zombie cells. The phenotypic switch will be controlled by an irreversible promoter flip accomplished through mutant CRE-Recombination LoxP sites. (Zhang 2002) This restructuring will result in a permanent change in genes being expressed upon infection.
In the above figure, the left cell depicts a Z-Mode or zombie cell. The main features of this are its expression of RFP and the expression of LasI, an enzyme that produces quorum signaling molecule acyl-homoserine lactone (AHL). The quorum molecule diffuses out of the zombie cell and into the normal cell. Upon AHL reaching high enough concentration, LasR will activate the expression of CRE and the genetic restructuring event may occur.
Mechanism of Infection
The mechanism of infection that the zombie cells use is based on a quorum sensing (QS) molecule produced by the LasI protein encoded for by the gene LasI from Pseudomonas aeruginosa. The QS molecule, acyl-homoserine lactone (AHL), permeates into the extracellular environment and is absorbed by the recipient normal cell. When AHL is in high enough concentration, the LasR enzyme from the gene LasR from Pseudomonas aeruginosa will convert AHL into a transcription factor to drive the LasR promoter, with the CRE-Recombinase protein under its control. CRE dictates the next behavior we want of our normal cells, to irreversibly switch into zombie cells, which can then infect other normal cells.
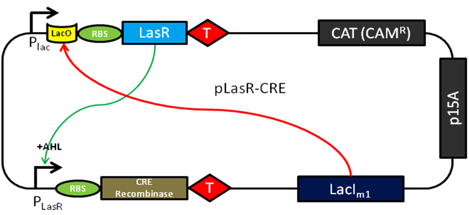
Features of the converter plasmid include a scheme for detecting the AHL quorum sensing molecule (from the LasR enhancer protein) and transduce the quorum molecule into the production of CRE recombinase. The expression of the LasR protein is under control of the endogenous Lac operator by the LacI protein. This feature will serve to limit the expression of this molecule during the cloning and testing of this parental plasmid sequence and allow swift induction of expression after adding IPTG. Upon the expression of the LasR protein and in the presence of a sufficient local concentration of AHL the CRE expression will be turned on.
Genetic Switch
Expression of CRE-Recombinase will then act as a genetic switch and trigger a restructuring in the Reporter plasmid. The genetic switch relies on a variant of the Cre/loxP recombination system. The protein CRE-Recombinase (causes recombination) catalyzes crossover events at specific DNA sequences termed loxP sites. Two variants, lox66 and lox71 can be combined to create the irreversible crossover event we want. CRE recombinase will trigger a one way recombination event of a LacUV5 promoter (Zhang 2002) causing it to switch and express the proteins it was previously facing away from. The LacUV5 flip is the essential difference between our normal and zombie cells. In a normal cell, IPTG induction will cause GFP expression, but if a cell has received the QS molecule AHL and expressed CRE-Recombinase, then the promoter will express the opposite set of genes. This results in a zombie cell expressing a RFP and the gene LasI, upon which it can now make the quorum molecule acyl-homoserine lactone, making it capable of further infecting proximal cells.
Testing
Control plasmids
pReporter.permaflipped
This control plasmid ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K734006 BBa_K734006]) is the “permaflipped” version of pReporter ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K734005 BBa_K734005]). Its sequence is identical to that of the reporter plasmid but has the promoter flipped, as if already acted upon by CRE-Recombinase. Using this control will show whether Z-Mode ("infected") cells actually express the RFP when CRE-Recombinase is assumed to work as intended, flipping the promoter region and generating one double-mutant and one wild-type loxP site. When transformed into cells and induced with IPTG, all cells should glow red if this control works correctly.
Control Converter Plasmid
To test the sensitivity and noise of the converter plasmid (pConverter) a control plasmid ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K734004 BBa_K734004]) with GFP in place of CRE was constructed. This allows it to be determined if GFP is expressed even in the absence of the quorum sensing molecule and how much quorum sensing molecule is needed to induce the LasR promoter and in turn GFP expression. The control plasmid was tested for GFP expression without IPTG or AHL molecule, with IPTG alone, with AHL alone, and with IPTG and AHL. Fluorescence was not detected under any of these testing conditions, and so far, it is unknown why.
Construction and Results
pConverter
The converter plasmid ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K734003 BBa_K734003]) operates by inducing the expression of CRE recombinase in the presence of the AHL quorum sensing molecule. CRE is placed downstream of the LasR promoter which is induced when bound by LasR protein and the AHL molecule. LasR is constitutively expressed in the presence of IPTG because it is promoted by the Lac promoter. To create this converter plasmid, each gene was independently amplified with overlapping regions to the genes adjacent to it, then several overlap PCR reactions where performed to obtain two final pieces, one of the pACYC backbone and one with the inserted genes, put together using a Gibson Assembly protocol. The same process was used to create the control converter plasmid. As a result of the inconclusive results of the control plasmid tests, the pConverter plasmid has not yet been tested.
pReporter
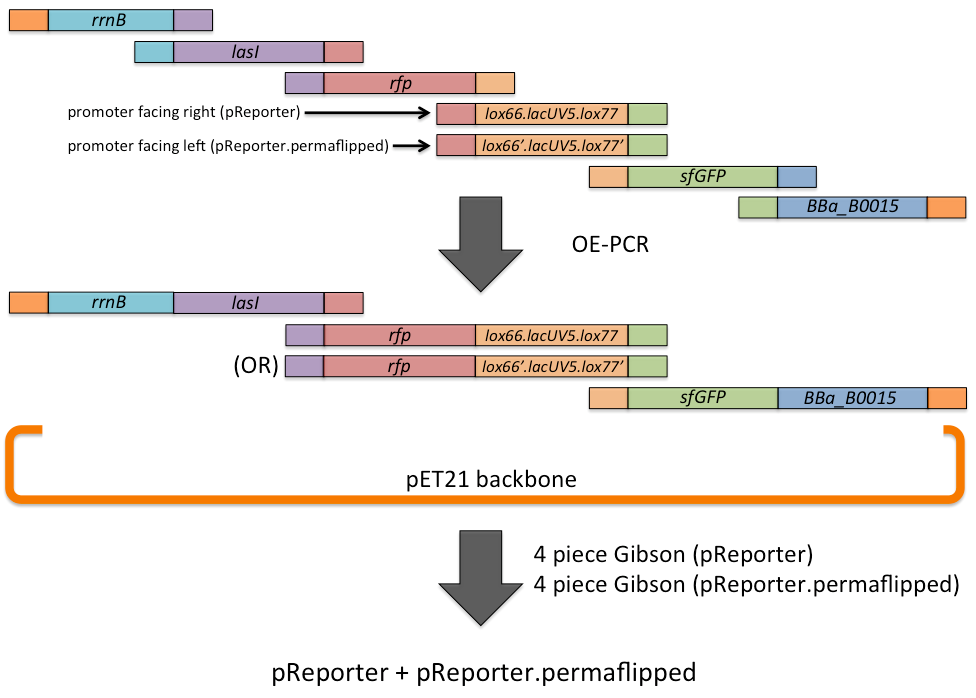
The pReporter plasmid and pReporter.permaflipped control were assembled via a combination of overlap-extension PCR and Gibson assembly. First, fragments (rrnB terminator, lasI, rfp, lox66.lacUV5.lox77 for pReporter and its corresponding permaflipped sequence, sfGFP, and the BBa_B0015 double terminator) were amplified with primers designed to add sequences to each side homologous to adjacent fragments. Then, OE-PCR was performed for each plasmid separately, resulting in a reduced number of pieces required for the subsequent Gibson reaction in order to increase efficiency. Gibson assembly resulted in the complete pReporter and pReporter.permaflipped plasmids.
A schematic is shown below.
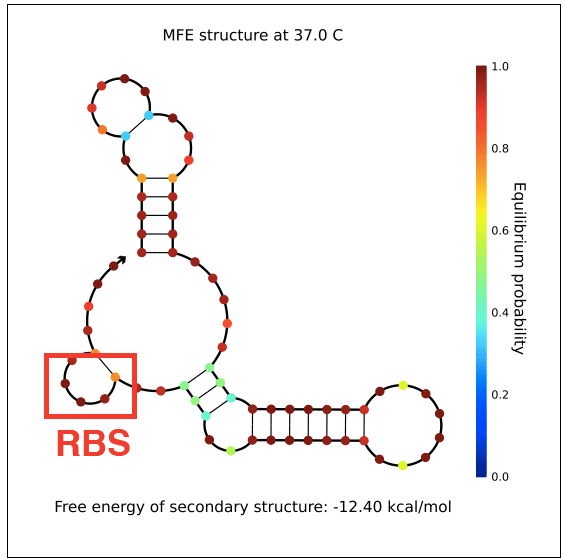
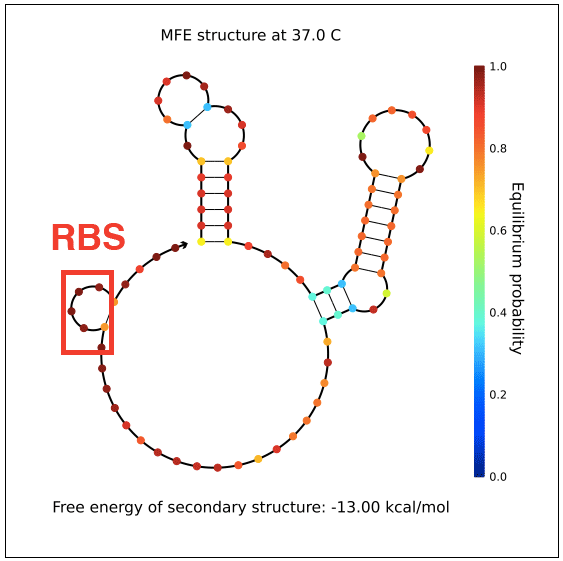
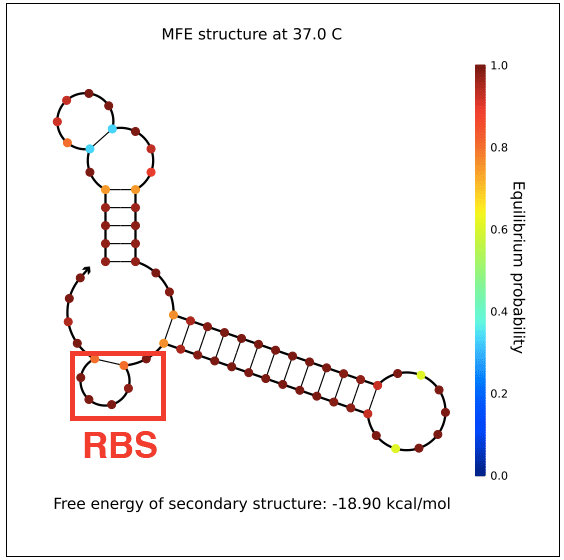
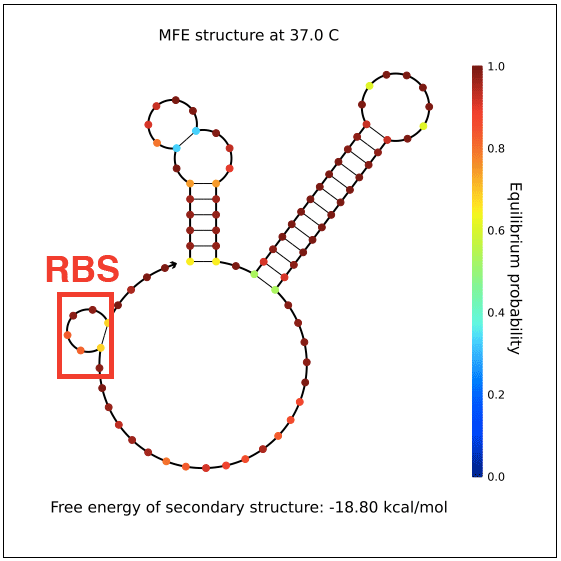
However, in testing, induction with IPTG does not allow GFP (in the case of pReporter) or RFP (in the case of either pReporter+constitutively expressed Cre or pReporter.permaflipped) expression, likely due to extensive secondary structure on the RNA transcripts. The loxP sites create hairpins that block the RBS. To address this problem, random nucleotide spacer regions could be inserted between the loxP sites and the RBSs of GFP and RFP. The table below shows [http://nupack.org NUPACK]-computed secondary structures with and without the proposed random nucleotide spacer regions.
 "
"