Team:TU Munich/Project/Vector Design
From 2012.igem.org



Contents |
Vector Design
What is the use of DNA sequences coding for valuable enzymes without the possibility to express them and analyze the protein activity?
As we planned to detect the enzymes from our biosynthetic pathways via Strep-tag II, a RFC25 compatible backbone was necessary. As no such backbone was availabe for yeast in the PartsRegistry, an important task at the beginning of our project was the design of an expression vector for yeast which is compatible to the iGEM cloning principles and standards. Based on the commercially available pYES2 vector, manufactured by Invitrogen, we created an vector containing:
- a RFC 25 compatible multiple cloning site
- a strep-tag II
- a 2µ origin for high copy number replication in Saccharomyces cerevisae
- the URA3 gene to use the uracil prototrophy of S. cerevisiae INVSc' as a selection marker for transfection
- a pUC origin for high copy number replication in E.coli
- the ß-lactamase coding gene to use ampicillin as a selection marker for cloning applications in E.coli
- a galacotsoe inducible PGal 1 promoter
- a CYC1 transcription terminator
Furthermore we designed several vectors containing our constitutive promoters Tef1, Tef2 and ADH and different additional terminators. The different versions of the vector have been successfully applied and tested in all other subprojects.
Design
The project work began with the replacement of the original multiple cloning side for a completely new designed multiple cloning site containing the typical RFC10/25 pre- and suffixes. Furthermore we integrated a gene sequence coding for a Strep-tag II in front of the suffix. This facilitates the purification and detection via Western Blot of expressed enzymes dramatically. The new multiple cloning side was constructed of four desoxyribooligonucleotides via oligonucleotide hybridization and ligation into the original pYES vector restricted with the outermost restriction enzymes of the old MCS.
In a further step all forbidden restriction sides of the enzymes used in the RFC10/25 standard in the vector backbone were deleted via side directed mutagenesis. The picture above shows different vector samples of this successive process which have been restricted with NgoMIV, PstI and SpeI: The resulting fragments decrease with each step leading to the pure vector backbone which is linearized by cutting in the multiple cloning side. Hence our first expression vector containing the original galactose inducible pGal 1 promoter was ready for cloning with iGEM standards!
A second step of this subproject was the exclusion of the f1 origin of replication for the phage λ and of the pGAL1 promoter. The resulting vector pTUM100 can used as powerful basis to integrate a wide variety of user defined promoters, genes and terminators. In our case we integrated the constitutive promoters Tef1, Tef2 and ADH in order to characterize them.
Results
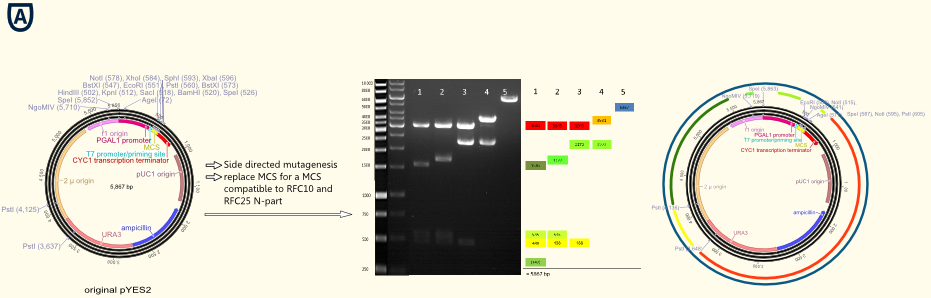
The figure on the right gives an opinion of all important elements located on the vector backbone:
- The T7 promoter primer binding site allows easy forward sequencing of integrated gene constructs using the standard T7 primer.
- The new RFC 10/25 compatible multiple cloning site enables straightforward cloning operations according to the iGEM standards.
- The strep-tag II serves as a powerful tool to purify and detect expressed proteins
- The CYC1 transcription terminator is an efficient and S. cerevisiae INVSc adjusted terminator of the transcription.
- The pUC origin permits high copy number replication in E.coli and therefore a simplified cloning.
- The ß-lactamase coding gene allows to use ampicillin as a selection marker for cloning applications in E.coli
- The URA3 gene renders possible to use the uracil prototrophy of the stem S. cerevisiae INVSc as a selection marker for transfection.
- The 2µ Origin of replication offers the opportunity of high copy number replication in S. cerevisiae INVSc
Based on the yeast shuttle vector without promoters called pTUM100 we created several vector versions containing constitutive promoters. This is represented in figures C to E:
- pTUM100 simply contains the new MCS, the transcription terminator and further elements required for cloning and transfection.
- pTUM102 to pTUM104 contain in addition the constitutive promoters pTef1, pTef2 and ADH.
- pTUM104 keeps the galactose inducible promoter pGAL1.
All of these versions were submitted as Biobricks.
BioBricks backbones
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801000 BBa_K801000] pTUM100 yeast shuttle vector without promoter
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801001 BBa_K801001] pTUM101 yeast shuttle vector with pTEF1 promoter
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801002 BBa_K801002] pTUM102 yeast shuttle vector with pTEF2 promoter
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801003 BBa_K801003] pTUM103 yeast shuttle vector with pADH1 promoter
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K801004 BBa_K801004] pTUM104 yeast shuttle vector with pGal 1 promoter
Induction assays with galactose
The pGAL1 promoter system is repressed in presence of glucose(West et al., 1984). Therefore maintaining cells in glucose containing medium causes the lowest basal transcription activity. Changing the carbon source to galactose de-represses the promoter so that transcription activity increases(Giniger et al., 1985). Alternatively cells can be maintained in raffinose which neither repress nor induce transcriptional activity. However the induction of transcriptional activity by galactose is accelerated by maintaining cells in raffinose. To characterize th pGAl1 promoter we cloned eGFP in a pTUM104 vector and transfected S. cerevisiae INVSccells. A single colony of 2-3 mm diameter was used to inoculate a overnight culture in 15 ml SC-U medium containing glucose. The OD at 600 nm was measured and an appropriate volume of cells was sedimented and resuspended in galactose containing SC-U media to create an initial OD of 0.4. In the further process samples of 1 ml volume were taken hourly and the OD at 600 nm as well as the fluorescence at 488 nm were determined. The fluorescence signal was normalized by divide by the OD600 signal.
 "
"
