Team:TU Darmstadt/Project/Transport
From 2012.igem.org
Transport
The objective of group "Transport" is the integration of a terephtalic acid uptake system in Escherichia coli (E. coli) . The uptake of TPA is crucial to produce high-value molecules into a E. coli. TPA can only pass the membrane at low pH values, which adversely affects the growth of E. coli. Therefore, a suitable transport system is needed that operates under optimal growth conditions for E. coli.
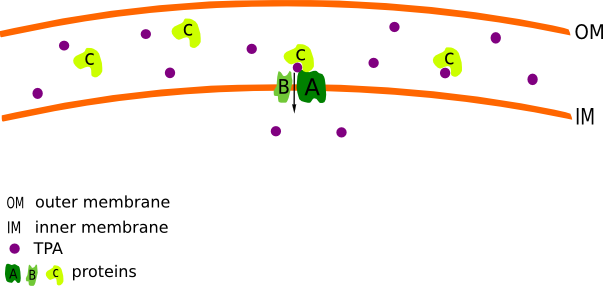
According to the data published by Sasoh et al.[1] Comamonas testosteroni (C. testosteroni) is able to utilizes TPA as the sole carbon and energy source. For that reason, we decided to isolate the putative TPA uptake System of C.testosteroni. This System bolongs to the tripartite tricarboxylate transporters and consists of three subunits (A to C). The large subunit A is a transmembrane protein with 11-12 alpha-helical spanners. It is acompanied by the small transmembrane subunit B which constis of 4-5 alpha-helical spanners. C is a specific periplasmic binding protein, which is moving freely in the periplasmic space and bonds to the AB units. (Fig.1) The function is similar to ABC transporters, however the sequences are unrelated. C. testosteroni features two different configurations of A and B proteins (A1 with 505 amino acids (aa) and B1 with 197 aa or A2 with 503 aa and B2 with 162 aa). The proteins are naturally unspecific and can transport different substrates. Initially A1B1C and A2B2C was isolated from C. testosteroni and be insert on pSB1C3 and pSB1A2 plasmides in E. coli DH5α. The Genes are regulated under the control of a Arabinose inducible promotor (araC-Pbad).
The intake of TPA is checked by photometry, gas chromatography-mass spectrometry (GC-MS) and energy dispersive X-ray spectroscopy (EDX). To determine the essential components and their combination for TPA transport into the cell, the genes were expressed in an overexpression strain like E.Coli C43(DE3). The structure characterisation was done by some bioinformatical tools like Protein Homology/anologY Recognition Engine V 2.0 (PHYRE2), I-TASSER servers, SignalIP 4.0 Server and TatP 1.0 Server.
Detailed information on our approach is available in the Transport Labjournal. For more informations concerning the other projects continue with 3. Metabolism.
[1] Sasoh, M., E. Masai, et al. (2006). "Characterization of the terephthalate degradation genes of Comamonas sp. strain E6." Appl Environ Microbiol 72(3): 1825-1832.
 "
"