Team:Wageningen UR/ObtainingthePoleroVLP
From 2012.igem.org
Isolation and BioBricking of the potato leaf roll virus (PLRV) coat protein.
Why work on PLRV?
Different viruses can be used to from VPL's. In our team we used both Hepatitis B and Cowpea Chlorotic Mottle Virus. But one can imagine that for different purposes of the VLP's, different characteristics of the VLP are desired. For example: the VLP should be stable at pH 7.4 when used for medical applications. For other applications the pH at which the VLP should be stable can be different. One could imagine that for various applications the desired size of the VLP can be different. In order to show that the production of VLP's can be done with a virus of choice we worked on Potato Leaf Roll Virus (PLRV). We isolated the Potato Leaf Roll Virus from infected potato plants and BioBricked the gene that encodes the viral coat protein. By creating a PLRV Coat Protein BioBrick we showed that it is possible to isolate a virus of your own choice, express the coat proteins in E.coli and form VLP's. This makes the PLRV CP BioBricks our favorite Natural BioBricks.
More about PLRV
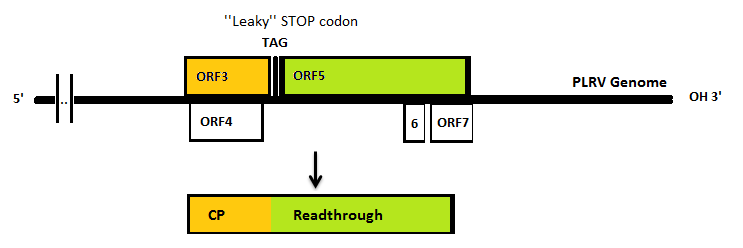
The Polero (potato leaf roll virus, PLRV) virus coat proteins can be used as building blocks to form VLP’s. PLRV is a positive sense RNA virus (group IV). Viruses from this group have their genome directly utilized as if it were mRNA. Ribosomes from the host cell translate their genome into a protein, with RNA-dependent RNA polymerase as one of it. This means that to isolate the coat protein gene we need to isolate the RNA of infected potato plants which would include the viral RNA.
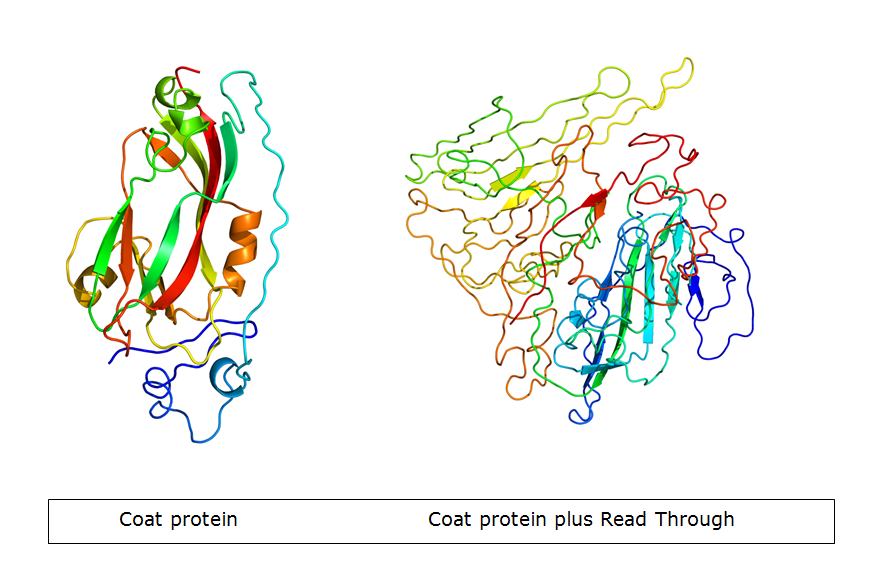
In nature, the stopcodon of the coat protein is sometimes ‘’missed’’, which results in a 70kDa read through product. This is considerably larger than the 23kDa coat protein on its own. When the coat protein as well as the coat protein plus read through are assembled together to form the virus capsid, spikes are formed on the capsid. These spikes, according to literature, have a function in the transmission by aphids, primarily the green peach aphid, Myzus persicae. We expect that these spikes can be changed without having a large effect on VLP formation. This, thus, seems a good strategy to build in either E-coils or K-coils, described in the general project introduction. Virus like particles (VLPs) are being formed either with or without read trough proteins {REFERENCE}. The expression of coat protein monomers, needed for VLP formation, has never been done in E.coli. If this step is successful, further modifications on the VLP’s will be done.
Work done on PLRV
After obtaining the coat protein gene of PLRV, the PLRV coat protein was sent to be sequenced. The sequence of our isolate together with other isolates described in literature gave us information about the conserved regions in the gene. Alignment experiments were performed, also together with data of other members of the Luteoviridae. The PLRV coat protein was thereafter ‘’BioBricked’’. This brick was expressed in Escherichia coli and produced monomers were isolated and purified.
Aim: Our primary aim is to obtain self-assembling VLP’s from PLRV in E. coli. If we succeed, we can attempt to modify the spike on the outside.
Progress:
- RNA isolation of potato leaf tissue
- Reverse transcriptase reaction to obtain cDNA
- PCR on cDNA using coat protein primers
- Addition of Prefix and Suffix to CP genes
- Transformation of the gene, inserted into iGEM pSB1C3 backbone, into E.coli
- Sequencing of coat protein gene
- Submission to Registry
- Luteoviridae coat protein/read trough analogy experiments
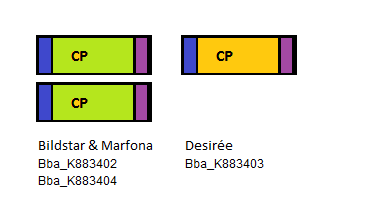
Four PLRV parts were sequenced: three PLRV coat proteins from different potato cultivars, one PLRV coat protein with a Histidine-tag added to the N-terminal. The three bare PLRV coat proteins were confirmed to be positioned nicely within the iGEM prefix and suffix. Unfortunately the sequence results showed the absence of the PstI restriction site in the suffix. The three different PLRV coat protein BioBrick parts were submitted to the Registry with the following accession numbers: BBa_K883402, BBa_K883403 and BBa_K883404.
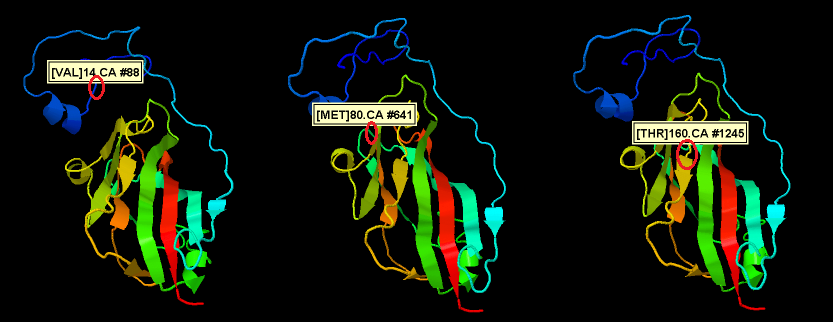
Alignment of the different PLRV isolates shows 9 SNP’s but not all SNP’s result in a different amino acid (only 3). The position of these amino acid changes if shown in the figure below.
Future work
- Isolation of monomers and formation of VLP’s
- VLP analysis/characterization
We inserted an IPTG inducible promotor upstream the PLRV CP gene. We performed 2 attempts to express the PLRV coat protein in E.coli but these were not successful. We obtained PLRV antibodies to use with Western blot to check for PLRV CP expression. Right now, we focus on getting the PLRV CP expressed. After successful expression we will try to isolate formed VLP's.
Further progress will be posted asap;)
 "
"