Team:Cambridge/Lab book/Week 10
From 2012.igem.org
(→Sunday (02/09/12)) |
(→Sunday (02/09/12)) |
||
| Line 132: | Line 132: | ||
*Gibson efficiency diagnostics | *Gibson efficiency diagnostics | ||
| + | |||
| + | ---- | ||
We decided to try to get to the bottom of why our Gibson assembly is so inefficient. Until this is fixed, we won't be able to do anything higher than 2-part reactions. | We decided to try to get to the bottom of why our Gibson assembly is so inefficient. Until this is fixed, we won't be able to do anything higher than 2-part reactions. | ||
Revision as of 16:17, 5 September 2012
| Week: | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|
Contents |
Monday (27/08/12)
Tuesday (28/08/12)
Wednesday (29/08/12)
Thursday (30/08/12)
PCR of Mg2+ riboswitch from genomic DNA
- Cycle settings:
- Melting - 98 °C - 10 seconds
- Annealing - 58 °C - 30 seconds
- Elongation - 72 °C - 30 seconds
- Fragments of correct size produced for all except lane 5 produced. In this lane, DNA appears to have accumulated in the well, indicating it may be genomic. In future, will use lower numbers of template cells to avoid getting so much genomic DNA.
- Products extracted and purified.
PCR of PJS130 vector for Mg riboswitch construct
- Cycle settings:
- Melting - 98 °C - 10 seconds
- Annealing - 60 °C - 30 seconds
- Elongation - 72 °C - 110 seconds
- Fragment of correct size produced in lane 5, but too faint to be extracted successfully. None of the other lanes were successful. We will try this again tomorrow, at 58 °C and with 35 cycles (as is usual) instead of 30.
- Note that phusion enzyme was left with primers and template DNA for about half an hour before reaction began, possibly causing degradation of the primer DNA.
Friday (31/08/12)
PCR of PJS130 vector for MgRS construct
- Cycle settings:
- Melting - 98 °C - 10 seconds
- Annealing - 58 °C - 30 seconds
- Elongation - 72 °C - 120 seconds
- Products of correct sizes (5.5kbp and 3.5 kbp) produced for all reactions, although lanes 2 and 4 failed to produce any product, despite primer smear. Most likely, template was not added, or one of the primers was not added.
- Products excised and purified.
Gibson assembly of Mg2+ riboswitch construct
- Reaction 1: Without 8 codon substitution: Vec A, Vec B -8 (replicate 1), Genomic -8.
- Reaction 2: Without 8 codon substitution: Vec A, Vec B -8 (replicate 2), Genomic -8.
- Reaction 3: With 8 codon substitution: Vec A, Vec B +8, Genomic +8 (replicate 1).
- Reaction 4: With 8 codon substitution: Vec A, Vec B +8, Genomic +8 (replicate 2).
Transformation of e.coli with Gibson products
- 20 μl of Gibson reaction mix transformed into e.coli cells. Transformants plated out onto 100 μg/ml ampicillin plates.
Saturday (01/09/12)
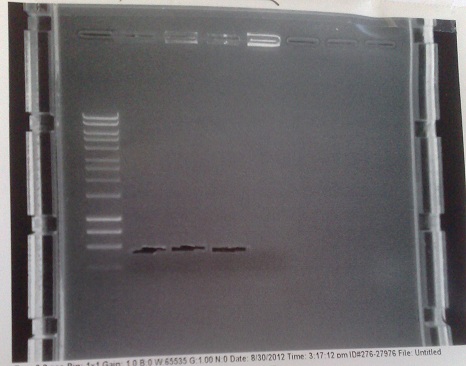
Verification of Mg2+ gel extractions
- DNA used in Gibson from yesterday and several weeks ago. Run on gel to verify the presence of DNA fragments of the correct size after gel extraction.
- Old DNA appears to have degraded, or else products were produced in too small a quantity to show up on the gel. Fortunately, these are not/ have not been used.
- Bands of the correct size for fragment A (5.5kbp), fragment B (3.5kbp) and the genomic DNA (550bp) present. Though riboswitch DNA has not come out well in this image, it was present under visual examination.
- Approximate DNA concentrations in the range of 2 - 6 ng/μl.
Sunday (02/09/12)
Verification of Mg2+ riboswitch construct by colony PCR
- Gibson primers used to amplify out riboswitch section of plasmid produced and transformed into e.coli to verify if
- E.coli from strains made yesterday
Verification of Mg2+ riboswitch construct by restriction digest
- Gibson efficiency diagnostics
We decided to try to get to the bottom of why our Gibson assembly is so inefficient. Until this is fixed, we won't be able to do anything higher than 2-part reactions.
PJ, a member of the haseloff lab whose gibson assemblies are working efficiently agreed to help us. Using our control fragments, PJ assembled them with our master mix and his master mix, and transformed them into his competent E.coli. Similarly, we assembled our fragments (from the same tube) with our master mix, but using his competent cells and our competent cells. This should show us if there is a problem with our mix or cells.
 "
"