Team:Cambridge/Lab book/Week 8
From 2012.igem.org
(Difference between revisions)
(→Saturday (18/08/12)) |
|||
| Line 121: | Line 121: | ||
*DNA extracted and purified. | *DNA extracted and purified. | ||
| - | '''[[Team:Cambridge/Protocols/ | + | '''[[Team:Cambridge/Protocols/PCRcolony|PCR of Mg2+ riboswitch construct vector]]''' |
---- | ---- | ||
| Line 135: | Line 135: | ||
:*Elongation - 72 °C - 100 seconds | :*Elongation - 72 °C - 100 seconds | ||
| - | * | + | *Products run on gel. No fragments produced. Positive control worked however, so master mix clearly works. |
| + | |||
| + | *Given this PCR has worked in the past, will try to re-run with the same settings and hope for success in the future. | ||
{{Template:Team:Cambridge/CAM_2012_TEMPLATE_FOOT}} | {{Template:Team:Cambridge/CAM_2012_TEMPLATE_FOOT}} | ||
Revision as of 16:41, 31 August 2012
| Week: | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|
Contents |
Monday (13/08/12)
Tuesday (14/08/12)
Gibson assembly of positive control
- Fragments used:
- Protocol changed slightly: 0.5 μl of each DNA fragment solution mixed in with master mix to make up 4 μl total volume (1μl DNA and 3μl master mix).
Transformation of e.coli with positive control DNA
- Chemically competent e.coli cells transformed with plasmid DNA produced by Gibson assembly step.
- Transformants plated out on 50 μg/ml kanomycin plates. Incubated overnight at 37 °C.
Wednesday (15/08/12)
PCR of positive control fragments
- Standard PCR settings used
Extraction of positive control DNA
- Positive control DNA from gel excised and purified.
- Additional elution steps used to concentrate resultant solution.
- Verified with nanodropper - final DNA concentrations:
Gibson assembly of positive control DNA
Transformation of positive control DNA into e.coli
- Chemically competent e.coli transformed with positive control Gibson products made previously.
- Transformants plated out onto 50 μg/ml kanomycin plates and incubated at 37 °C overnight.
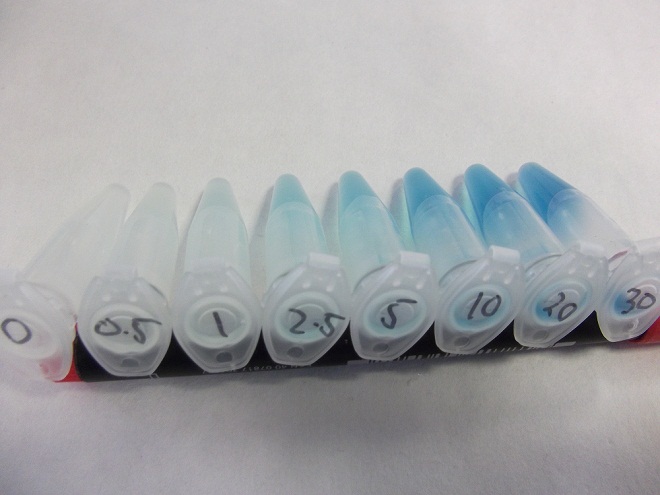
Characterization of fluoride riboswitch construct
- Strain of bacillus lacking fluoride transporter system tested. Concentrations used:
- 0mM - 0.5mM - 1mM - 2.5mM - 5mM - 10mM - 20mM - 30mM
Thursday (16/08/12)
Characterization of fluoride riboswitch construct
- Eppindorfs containing the bacillus with the fluoride riboswitch removed from incubator and imaged.
- Now this definitely works, we will try quantifying this riboswitch with an ONPG assay and the plate reader.
PCR ran by paul - insert photos/discuss
- Only positive control and two sets of triplicates worked
- YFP and CFP extracted successfully
Friday (17/08/12)
- Team meet up today! No lab work was done.
Saturday (18/08/12)
PCR of Fluoride biobrick format
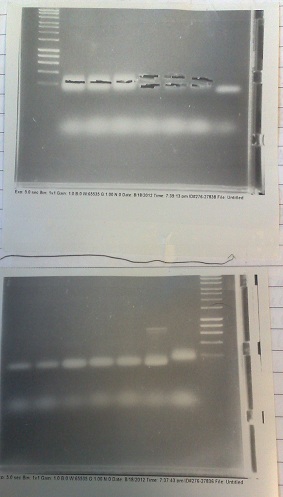
Gels from PCR of Fluoride BB and PJS130 fragments. Top: Lanes 2-4: Gibson compatable Fluoride biobrick fragment. Lanes 5-7: Ligation compatable Fluoride biobrick fragment. Lane 8: PJS130 fragment A. Bottom: Lanes 1-2: PJS130 fragment A. Lanes 3-5: PJS 130 fragment B. Lane 6: Positive control. Lane 7: Negative control.
- Two sets of primers used: one to allow insertion into backbone by ligation, and one to allow insertion by Gibson assembly.
- Cycle settings:
- Melting - 98 °C - 10 seconds
- Annealing - 58 °C - 30 seconds
- Elongation - 72 °C - 100 seconds
- Products run on gel. Fragments produced of correct size, however they overlapped with the primer dimer band due to the small size of the fragments.
- DNA extracted and purified.
PCR of Mg2+ riboswitch construct vector
- Separate reactions for
- Cycle settings:
- Melting - 98 °C - 10 seconds
- Annealing - 58 °C - 30 seconds
- Elongation - 72 °C - 100 seconds
- Products run on gel. No fragments produced. Positive control worked however, so master mix clearly works.
- Given this PCR has worked in the past, will try to re-run with the same settings and hope for success in the future.
 "
"