Team:TU Munich/Project/Overview
From 2012.igem.org
VolkerMorath (Talk | contribs) (→pTUM100) |
VolkerMorath (Talk | contribs) (→pTUM100) |
||
| Line 127: | Line 127: | ||
All BioBricks were sequenced. Sequences can be found in the registry of standard biological parts under the following entries: | All BioBricks were sequenced. Sequences can be found in the registry of standard biological parts under the following entries: | ||
| - | + | pTUM100 (<partinfo>BBa_K801000</partinfo>), pTUM101 (<partinfo>BBa_K801001</partinfo>), pTUM102 (<partinfo>BBa_K801002</partinfo>), pTUM103 (<partinfo>BBa_K801003</partinfo>) and pTUM104 (<partinfo>BBa_K801004</partinfo>). | |
Revision as of 15:01, 26 October 2012



Contents |
Overview
Vision
We, TU Munich’s 2012 iGEM team, strive to catalyze the diffusion process of knowledge about genetic engineering and synthetic biology among the general public. Using the example of iGEM’s first and finest SynBio Beer we involve, interest and inspire people to reconsider preconceived ideas and encourage them to openly engage in a broad discussion weighing pros and cons of genetic engineering in foodstuff. We sketch a future where new technology can be applied in a meaningful way to complement traditional foods or beverages.
Biosynthesis pathways
Limonene
Limonene is a cyclic terpene and a major constituent of several citrus oils. D-Limonene has been used as a component of flavorings and fragrances. It is formed from geranyl pyrophosphate by limonene synthase.
We successfully demonstrated the production of the flavoring substance limonene by expressing limonene synthase in S. cerevisiae, which naturally synthesizes the educt geranyl pyrophosphate.
(+)-Limonene synthase 1 http://partsregistry.org/Part:BBa_K801065 BBa_K801065 and (+)-limonene synthase 1 with yeast consensus sequence http://partsregistry.org/Part:BBa_K801060 BBa_K801060 were successfully cloned into our new yeast expression vector pTUM100. Expression of recombinant limonene synthase in Saccharomyces cerevisiae was proven by western blotting. Subsequently the protein was purified using SA-chromatography and size exclusion chromatography. The functionality of the enzyme was verified by in vivo and in vitro detection of limonene via GC-MS.
Furthermore, we established gene constructs of the limonene synthase coding sequence with different yeast specific promoters and terminators http://partsregistry.org/Part:BBa_K801062 BBa_K801062, http://partsregistry.org/Part:BBa_K801063 BBa_K801063, http://partsregistry.org/Part:BBa_K801064 BBa_K801064.
Last but not least, we have brewed iGEM's first SynBio beer containing limonene.
We have achieved functional expression of Citrus limon limonene synthase and production of limonene in yeast. The last remaining step to a SynBio beer would be the integration of our gene constructs http://partsregistry.org/Part:BBa_K801062 BBa_K801062, http://partsregistry.org/Part:BBa_K801063 BBa_K801063, http://partsregistry.org/Part:BBa_K801064 BBa_K801064 into the yeast genome.
Thaumatin
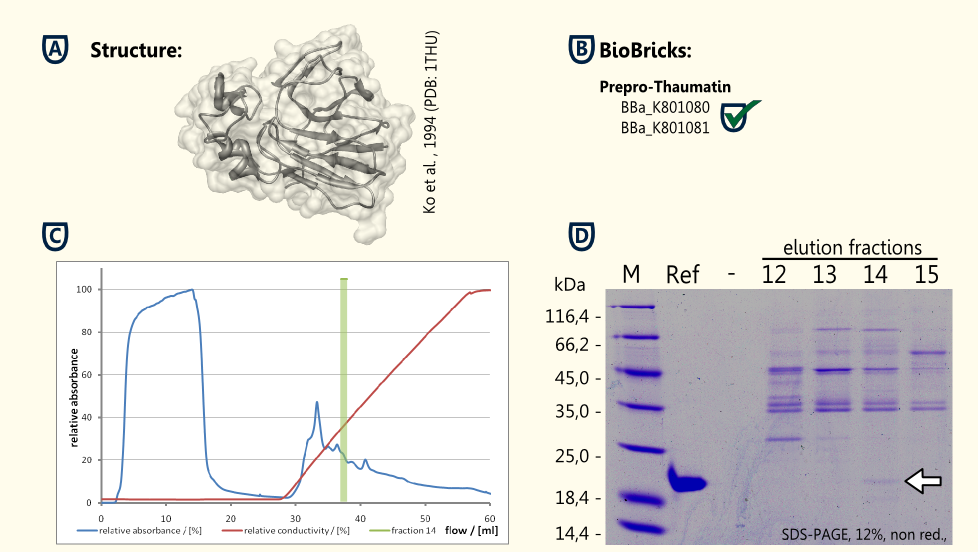
Thaumatin is a natural α+β-protein which is synthesized by the katamfe plant (Thaumatococcus daniellii). It is said to be 2,000 to 100,000 times sweeter than sucrose on molar basis, but the sweetness builds up slow and lasts long. It has been approved as a sweetener by the European Union (E957).
Our aim is to have S. cerevisiae secrete functional thaumatin by expressing preprothaumatin – a principle which has been proven by http://www.ncbi.nlm.nih.gov/pubmed/6327079 Edens et al., 1984.
The BioBrick for preprothaumatin [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801080 BBa_K801080] as well as an expression cassette [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801080 K801080] were successfully cloned, expressed in yeast, purified using an ion exchange chromatography (see figure C) and detected in the SDS-PAGE. Therefore, the expression of thaumatin in yeast could be demonstrated and functionality of the BioBrick is confirmed.
A proof of principle for the expression of thaumatin was achieved. Further goals are the increase of the expression of thaumatin and the investigation of the secretion.
Caffeine
Caffeine is a purine-alkaloid and its biosynthesis is known from coffee plants and tea plants. The molecule acts as a competitive antagonist of adenosine receptors and, therefore, increases indirectly neurotransmitter concentrations resulting in warding off drowsiness and restoring alertness.
The idea is to perform a heterologous gene expression of the three enzymes 7-methylxanthosine synthase (CaXMT1), N-methyl nucleosidase (CaMXMT1) and caffeine synthase (CaDXMT1) required for caffeine biosynthesis in Saccharomyces cerevisiae.
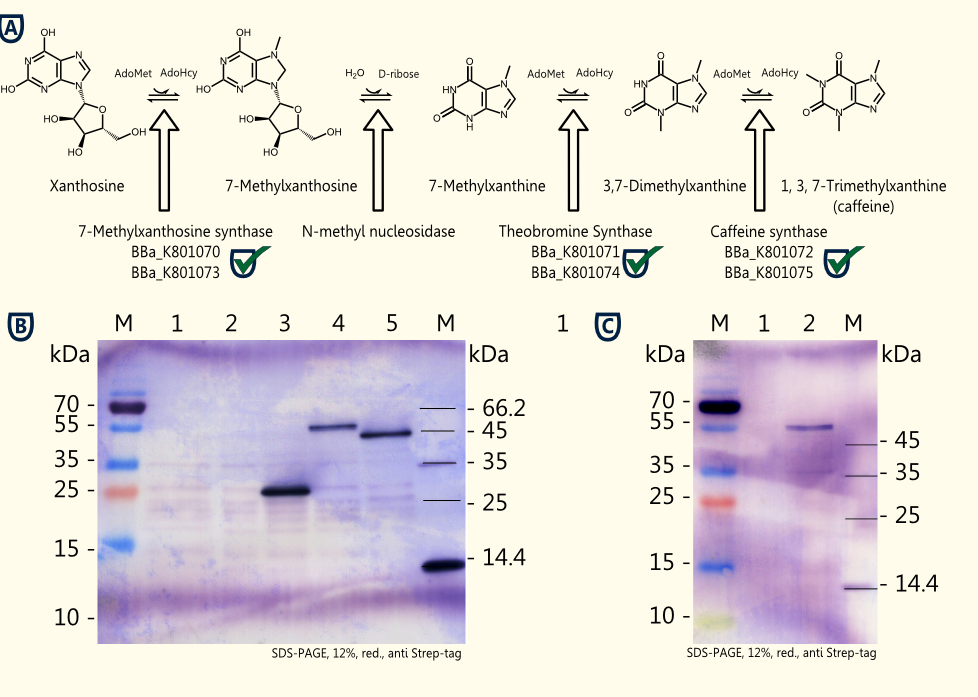
- Successful cloning of the three enzymes [http://partsregistry.org/Part:BBa_K801070 7-methylxanthosine synthase (CaXMT1)], [http://partsregistry.org/Part:BBa_K801071 N-methyl nucleosidase (CaMXMT1)] and the [http://partsregistry.org/Part:BBa_K801072 caffeine synthase (CaDXMT1)] into the shuttle vector pTUM104 and pSB1C3 each.
- Successful assembly of the BioBricks to form expression cassettes consisting of promoter, gene and terminator: [http://partsregistry.org/Part:BBa_K801073 pTEF2-CaXMT1-tADH1], [http://partsregistry.org/Part:BBa_K801074 pTEF1-CaMXMT1-tADH1] and [http://partsregistry.org/Part:BBa_K801075 pTEF2-CaDXMT1-tADH1]) into pSB1C3.
- Successful assembly of the expression cassettes of the three relevant enzymes forming a composite part of 6.4 kb capable of caffeine production in yeast ([http://partsregistry.org/Part:BBa_K801077 Caffeine Synthesis Pathway]) into pSB1C3.
- Successful expression of CaXMT1, CaMXMT1 and CaDXMT1 in Saccharomyces cerevisiae INVSc1 in selective Sc minimal induction medium lacking uracil with 2 % galactose.
The homologue expression of the three required enzymes for caffeine synthesis in Saccharomyces cerevisiae INVSc1 transformed with pTUM102_CaXMT1, pTUM102_CaMXMT1 and pTUM102_CaDXMT1 was successful. Further testing of caffeine production using crude extracts from lysed transformed yeast cells and working on genome integration of the expression cassette containing all three enzymes flanked by promoter and terminator are in progress.
Xanthohumol
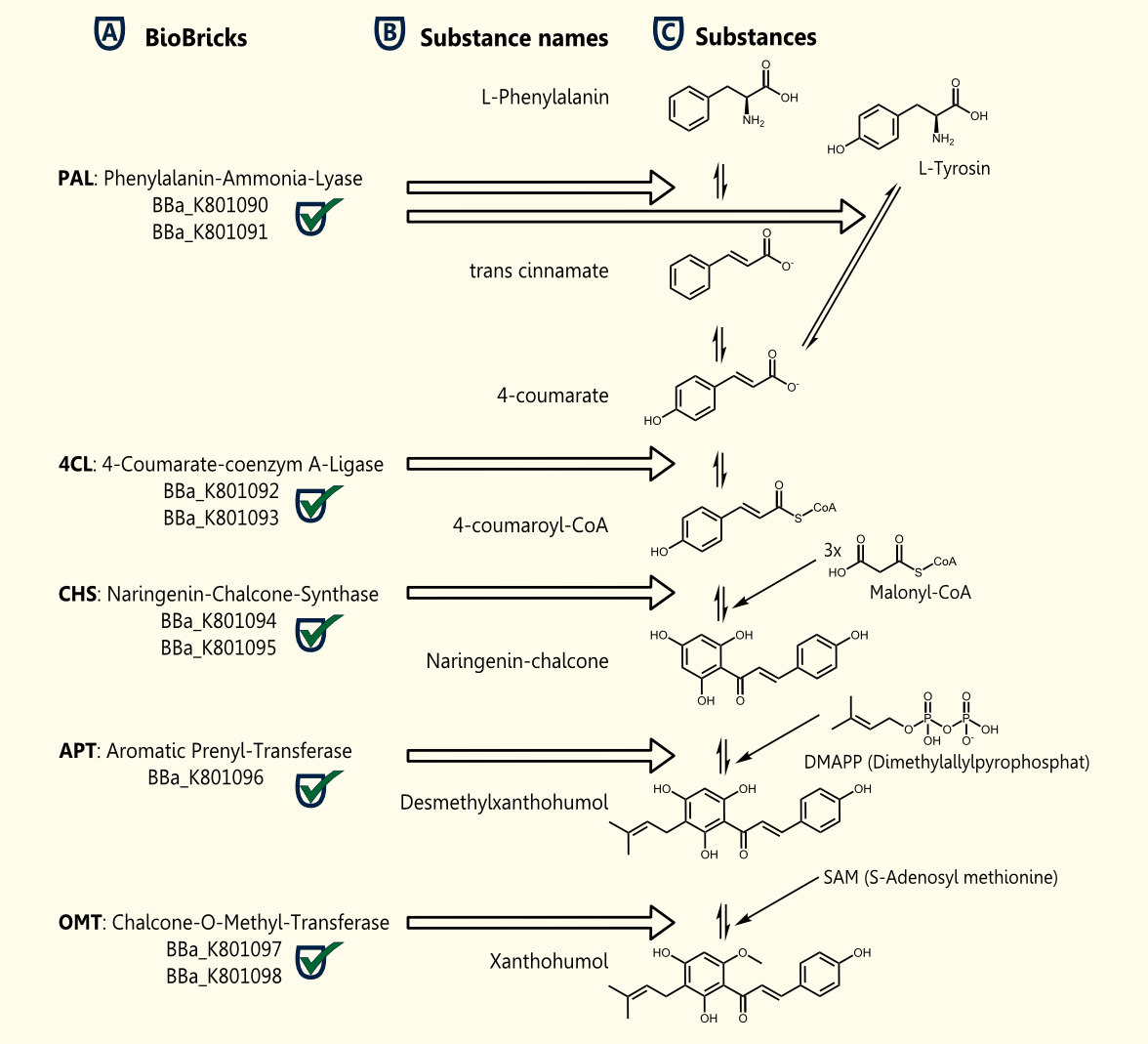
Xanthohumol is known as a putative cancer chemopreventive agent due to its antioxidant activities http://www.ncbi.nlm.nih.gov/pubmed/10995285 Miranda et al., 2000. Our goal is a heterologous gene expression of all enzymes required for xanthohumol biosynthesis in S. cerevisiae.
The pathway for the production of this plant secondary metabolite is composed of five steps, starting with the conversion of phenylalanine and followed by four further enzymatic reactions.
The whole biosynthetic pathway for the production of xanthohumol was converted into BioBricks. Except for APT each of the enzymes were cloned in two versions one having the proposed consensus sequence for more efficient expression in yeast chassis and another for usage of these BioBricks in other chassis. All BioBricks were sequenced. Sequences can be found in the registry of standard biological parts: [http://partsregistry.org/Part:BBa_K801090 BBa_K801090], [http://partsregistry.org/Part:BBa_K801091 BBa_K801091], [http://partsregistry.org/Part:BBa_K801092 BBa_K801092], [http://partsregistry.org/Part:BBa_K801093 BBa_K801093], [http://partsregistry.org/Part:BBa_K801094 BBa_K801094], [http://partsregistry.org/Part:BBa_K801095 BBa_K801095], [http://partsregistry.org/Part:BBa_K801096 BBa_K801096], [http://partsregistry.org/Part:BBa_K801097 BBa_K801097] and [http://partsregistry.org/Part:BBa_K801098 BBa_K801098]
The construction of the xanthohumol pathway was achieved, whereas the expression and characterization have to be postponed after the European Jamboree or might be an interesting task for next year's iGEM teams.
Vector Design
pTUM100
Designing an expression vector for yeast which is compatible to the iGEM cloning principles and standards was the main aim of this subproject. Based on the commercially available pYES2 vector we created vectors containing inducible and constitutive promoters in order to establish efficient possibilities to clone and express our enzymes.

Using the pYES vector from Invitrogen we first deleted five forbidden restriction sites in the vector backbone via side directed mutagenesis. Furthermore, the original multiple cloning site was replaced by a multiple cloning site compatible with the RFC 10/25 cloning standards. To allow easy extraction and purification of proteins for in vitro applications the new multiple cloning site allows to express proteins with a Strep-tag II. Exclusion of the galactose inducible promoter provided a powerful basis vector for the integration of user-defined promoters. This way the pTUM100 vector gives a valuable contribution to our and to further protein expression and promoter characterization experiments in Saccharomyces cerevisiae. Moreover, we used the pTUM100 to integrate the three constitutive promoters Tef1, Tef2 and ADH which come all with different promoter intensities.
The galactose inducible expression system was a great aid for the majority of all subprojects. Especially the opportunity to purify and detect (via Western blot) proteins using the Strep-tag II did facilitate our laboratory practice and accelerated our work progress. To cover even more demands we are planning to design a second vector template containing a His-tag.
All BioBricks were sequenced. Sequences can be found in the registry of standard biological parts under the following entries:
pTUM100 (<partinfo>BBa_K801000</partinfo>), pTUM101 (<partinfo>BBa_K801001</partinfo>), pTUM102 (<partinfo>BBa_K801002</partinfo>), pTUM103 (<partinfo>BBa_K801003</partinfo>) and pTUM104 (<partinfo>BBa_K801004</partinfo>).
Regulation of Genexpression
By developing inducible promoters and placing them upstream of our biosynthetic pathways we create the possibility to make S. cerevisiae dynamically respond to concentration changes in its medium as well as to external stimuli.
An optimal inducing substance needs to be inexpensive, nontoxic and fully controllable in its application. Only substances with these characteristics allow to precisely regulate a system temporally, spatially and quantitatively.
Ethanol-inducible promoter
The KlADH4-promoter from the yeast Kluyveromyces lactis regulates the expression of a mitochondrial alcohol dehydrogenase in an ethanol-dependent way. An alcohol-inducible promoter would be incredibly useful for anyone planning to brew a beer with a transgenic yeast - it would allow for the induction of the target genes after the main fermentation has finished and this way, the metabolic burden for the yeast cells could be lowered. All the transcription factors known to be involved in the regulation of the KlADH4-promoter in K. lactis also occur in S. cerevisiae http://www.ncbi.nlm.nih.gov/pubmed/10724480 Mazzoni et al., 2000. This is why we are confident that this promoter maintains its unique characteristics when transformed into S. cerevisiae.
At this time our results concerning the KlADH4-promoter (originally from the yeast Kluyveromyces lactis) suggest that this promoter is ethanol inducible in S. cerevisiae. Further experiments are still being done to abolish residual ambiguities. The lowest ethanol concentration at which eGFP-expression was detected is 0.9 Vol.-%.
Because S. cerevisiae is such a good brewer, it was difficult to produce a stringent negative control in our characterization experiments. However, we finally figured out some experiments that allowed us to keep the ethanol concentration below 0.5 % v/v, which is a concentration at which induction is observed in K. lactis. We are working hard on providing additional data, but we are confident that we will be able to provide clear evidence that this promoter is ethanol-inducible not only in K. lactis, but also in S. cerevisiae.
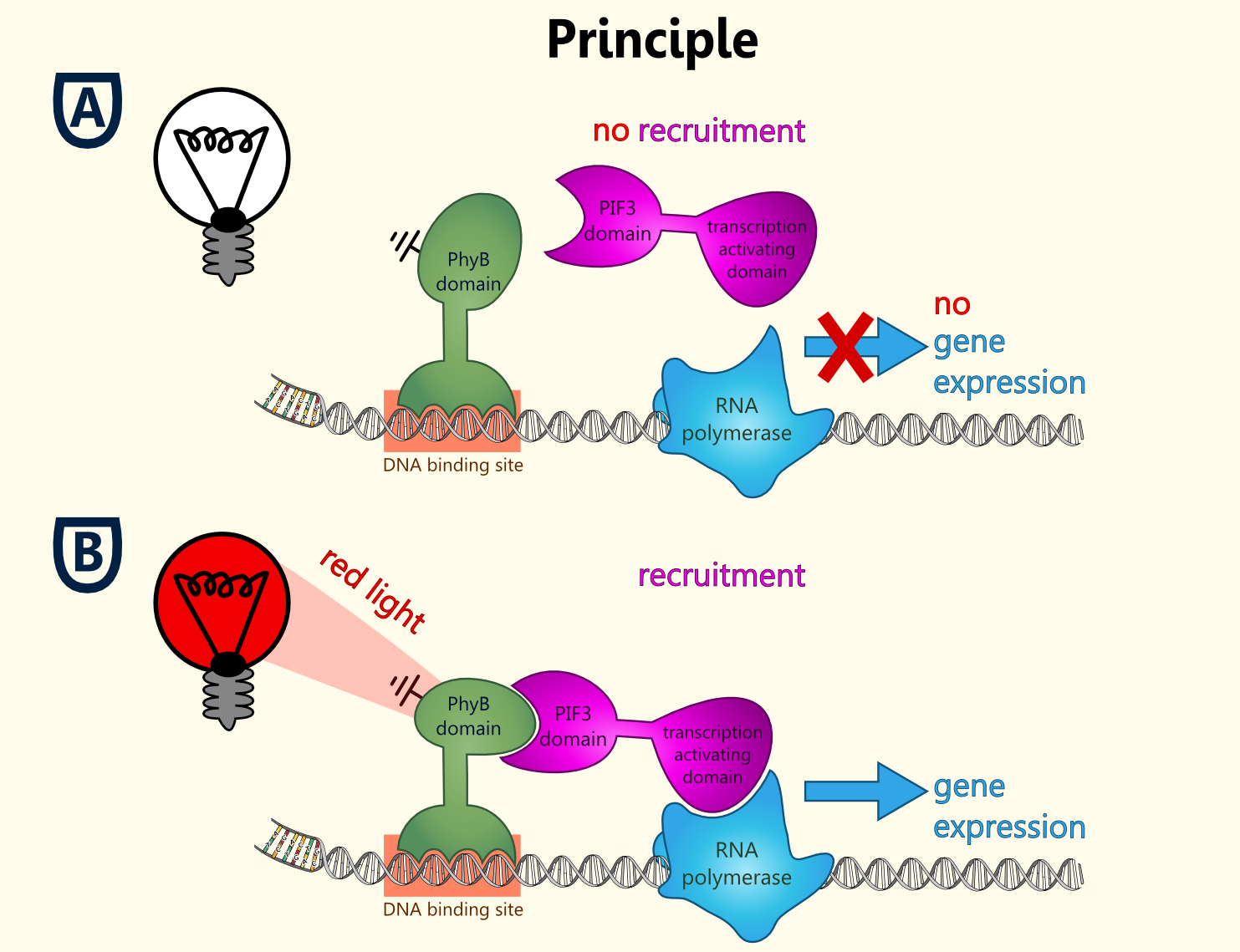
Light-switchable promoter
The idea behind a light-switchable system is to create a gene expression system which can be induced and deactivated by light of a certain wavelength.
This system is extremely attractive, as induction does not require the addition of a specific substance. This makes induction cheap, fast, precise and also compatible with the Bavarian purity law.
All fusion proteins for the two types of a light-switchable promoter system has been finished ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K801039 BBa_K801039], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801040 BBa_K801040] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801041 BBa_K801041]), also gene expression batteries coding for all components of each type of our light-switchable promoter system has been done ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K801042 BBa_K801042] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801043 BBa_K801043]). Since lacking of a second functional yeast vector carrying another auxotrophy marker than URA3 of the pTUM plasmids, which is already reserved for the biosynthesis enzymes, proteins and also reporters, we were not able to clone the whole gene expression battery, into a yeast vector, in order to co-transfect the yeast with one plasmid with the reporter construct and the second plasmid coding for all the devices needed in a light-switchable promoter system.
To get gene expression casette for both of the light-switchable promoter systems ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K801042 BBa_K801042] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801043 BBa_K801043]) into a yeast plasmid, we want to use pSB6A0 ([http://partsregistry.org/Part:BBa_K268000 BBa_K268000]) carrying a TRP1 auxotrophy marker. But we've discovered some discrepancies in this part, e. g. there is still at least one forbidden restriction site (XbaI or PstI, please see experience site of the part). So we have to correct the part by quickchange mutagenesis.
After successful cloning and transfection of the gene expression batteries including their reporters, we will proceed with the characterization of the part.
In parallel, we will implement also the whole biosynthesis pathway for the phycocyanobilin synthesis, downstream of our both batteries for a light-switchable promoter system ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K801042 BBa_K801042] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K801043 BBa_K801043]); after that, we don't have to add phycocyanobilin in our medium anymore!
If there is still time in the end, we will clone our system including the biosynthetic genes for PCB synthesis in the yeast integrative vector ([http://partsregistry.org/Part:BBa_K300001 BBa_K300001]) and will start a genome integration in order to create a new yeast strain with a fully functional light-switchable promoter system.
Genome Integration
As we cannot obey the letter of the German purity law (there is a zero tolerance policy concerning transgenic ingredients), we try our best to meet the spirit. Thus, it is unacceptable for us to work with antibiotics to keep up the selective environment. Since we cannot work with auxotrophies in beer either, we have to make sure the yeasts do not lose the BioBricks. The most promising way to accomplish a long lasting presence of our constructs is to achieve genome integration.
First experiments to characterize the function of the yeast integration system were performed and the used selection marker was maintained in the yeast culture, although the selection pressure was switched off. This indicates that first integrations were achieved.
Maintaining the plasmids harboring our expression cassettes in the yeast cells during the brewing process is best possible using genome integration. This becomes increasingly interesting, when a yeast strain with different expression cassettes is to be created. Because this is intended for the next step of our project the integration of our expression cassettes becomes increasingly important.
Brewing our SynBio Beer
Contrary to popular opinion the chief ingredient of beer is not YPD but gyle, a carefully prepared mixture of malt, hop and water. Although the name of the yeast strain commonly used in the lab, S. cerevisiae, suggests that it is used in the beer brewing process, the yeast strains generally employed in brewing process have strongly adapted to gyle, as they are reutilized in every succeeding brewing cycle. Hence some investigation on how our yeast performs in gyle was necessary.
Our experiments show that the growth of several different yeast strains is not impaired in gyle!
Expression assays proved the necessity of genome integration for a proper SynBio Beer.
As soon as cloning of integration vectors containing expression cassettes for our biosynthetic pathways are finished we will repeat the brewing experiments.
References
- http://www.ncbi.nlm.nih.gov/pubmed/6327079 Edens et al., 1984 Edens, L., Bom, I., Ledeboer, A. M., Maat, J., Toonen, M. Y., Visser, C., and Verrips, C. T. (1984). Synthesis and processing of the plant protein thaumatin in yeast. Cell, 37(2):629–33.
- http://www.ncbi.nlm.nih.gov/pubmed/10995285 Miranda et al., 2000 Miranda, C. L., Stevens, J. F., Ivanov, V., McCall, M., Frei, B., Deinzer, M. L., and Buhler, D. R. (2000). Antioxidant and prooxidant actions of prenylated and nonprenylated chalcones and flavanones in vitro. J Agric Food Chem, 48(9):3876–84.
- http://www.ncbi.nlm.nih.gov/pubmed/10724480 Mazzoni et al., 2000 Mazzoni, C., Santori, F., Saliola, M., and Falcone, C. (2000). Molecular analysis of uas(e), a cis element containing stress response elements responsible for ethanol induction of the kladh4 gene of kluyveromyces lactis. Res Microbiol, 151(1):19–28.
 "
"