Team:Peking/Modeling
From 2012.igem.org
Nathan 1991 (Talk | contribs) m |
|||
| Line 20: | Line 20: | ||
<p> | <p> | ||
Our <i>Luminesensor</i> is a ultra-sensitive fusion protein to sense 450nm to 470nm light and then regulate the gene expression.(<a href="/Team:Peking/Project/Luminesensor/Future#FigS">spectrum data here</a>) Although <i>Luminesensor</i> excels and eclipses similar systems due to its ultra-sensitivity and dynamic range, there are still several imperfect aspects. For example, the response time of the protein can be up to hours<sup><a href="#ref1" title="Zoltowski, B.D., Crane, B.R.(2008). Light Activation of the LOV Protein Vivid Generates a Rapidly Exchanging Dimer. Biochemistry, 47: 7012: 7019">[1]</a></sup> and the contrast of binding efficiency with and without light has much room for improvement. After modeling the DNA binding process of <i>Luminesensor</i>, we managed to find out four key parameters, two of which mainly control the response time, and the others control the contrast of binding efficiency. According to the feasibility in experiment, we tuned two most facile parameters of each aspect to optimize the performance of <i>Luminsensor</i>, and later figured out mutation sites related to these two parameters experimentally.<br /><br /> | Our <i>Luminesensor</i> is a ultra-sensitive fusion protein to sense 450nm to 470nm light and then regulate the gene expression.(<a href="/Team:Peking/Project/Luminesensor/Future#FigS">spectrum data here</a>) Although <i>Luminesensor</i> excels and eclipses similar systems due to its ultra-sensitivity and dynamic range, there are still several imperfect aspects. For example, the response time of the protein can be up to hours<sup><a href="#ref1" title="Zoltowski, B.D., Crane, B.R.(2008). Light Activation of the LOV Protein Vivid Generates a Rapidly Exchanging Dimer. Biochemistry, 47: 7012: 7019">[1]</a></sup> and the contrast of binding efficiency with and without light has much room for improvement. After modeling the DNA binding process of <i>Luminesensor</i>, we managed to find out four key parameters, two of which mainly control the response time, and the others control the contrast of binding efficiency. According to the feasibility in experiment, we tuned two most facile parameters of each aspect to optimize the performance of <i>Luminsensor</i>, and later figured out mutation sites related to these two parameters experimentally.<br /><br /> | ||
| - | Furthermore, in order to extend the application of <i>Luminsensor</i>, we managed to expand the spectrum of <i>Luminsensor</i> through model. Here we use | + | Furthermore, in order to extend the application of <i>Luminsensor</i>, we managed to expand the spectrum of <i>Luminsensor</i> through model. Here we use molecular modeling to identify the possiblity of modifications to the conjugating substructure of Flavin Adanine Dinucleotide (FAD) substrate, which acts as the chromophore of VVD. |
</p> | </p> | ||
</div> | </div> | ||
Revision as of 02:16, 27 September 2012
Summary
Our Modeling group conducted modeling to guide the optimization of our Luminesensor, combining protein kinetics, thermodynamics, and stochastic simulation with molecular docking methods together. Thereby the mutagenesis is indeed rationally guided by our modeling prediction, and experimental results excellently correspond to the model! We also used Molecular Modeling to expand the spectrum of Luminesensor, which would greatly extend its application. Additionally, we simulated the process of Phototaxis both on the macro level -- Mean-field PDE (partial difference equations) and on the micro level -- Stochastic Simulation. In the PDE model, we developed a hexagonal-coordinate simulation environment for dynamic system on a continuous plane, which would be a very useful prototype for future simulation on cellular movement, pattern formation and other potential systems.
Luminesensor Modeling
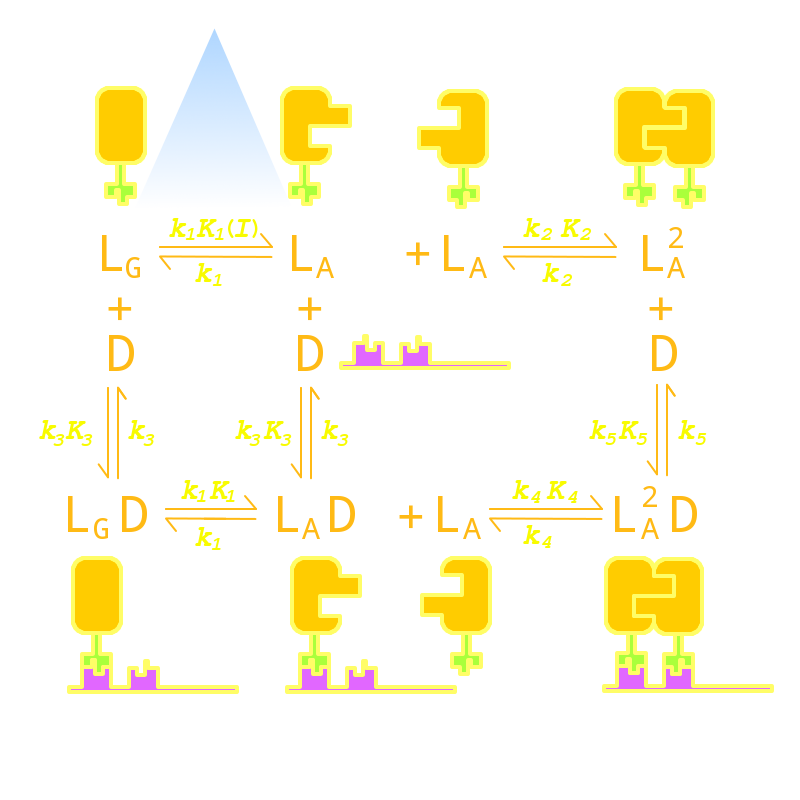
Our Luminesensor is a ultra-sensitive fusion protein to sense 450nm to 470nm light and then regulate the gene expression.(spectrum data here) Although Luminesensor excels and eclipses similar systems due to its ultra-sensitivity and dynamic range, there are still several imperfect aspects. For example, the response time of the protein can be up to hours[1] and the contrast of binding efficiency with and without light has much room for improvement. After modeling the DNA binding process of Luminesensor, we managed to find out four key parameters, two of which mainly control the response time, and the others control the contrast of binding efficiency. According to the feasibility in experiment, we tuned two most facile parameters of each aspect to optimize the performance of Luminsensor, and later figured out mutation sites related to these two parameters experimentally.
Furthermore, in order to extend the application of Luminsensor, we managed to expand the spectrum of Luminsensor through model. Here we use molecular modeling to identify the possiblity of modifications to the conjugating substructure of Flavin Adanine Dinucleotide (FAD) substrate, which acts as the chromophore of VVD.
Phototaxis Modeling
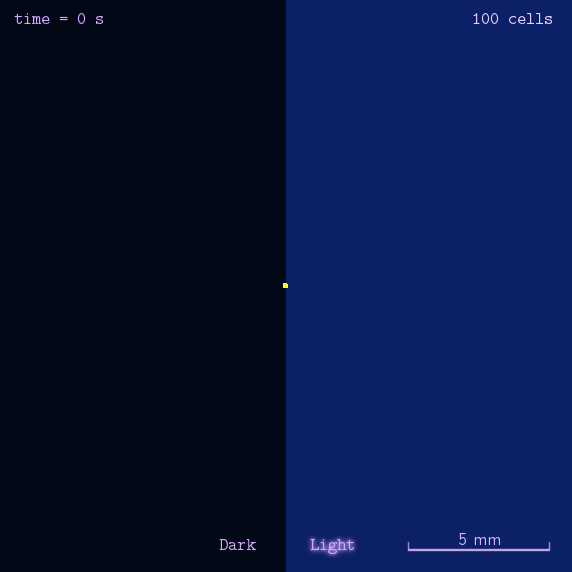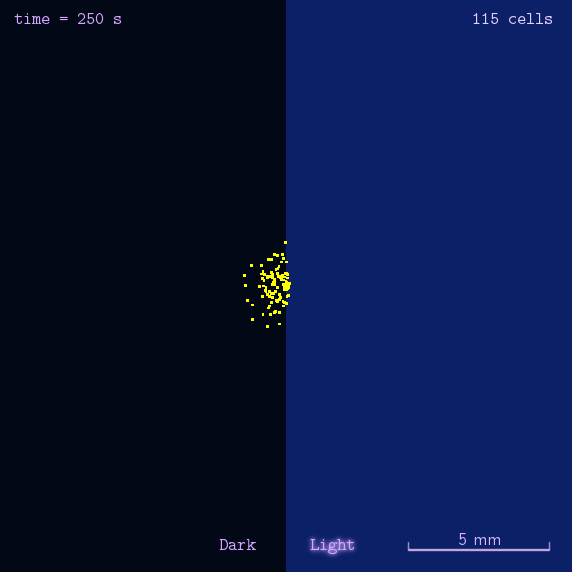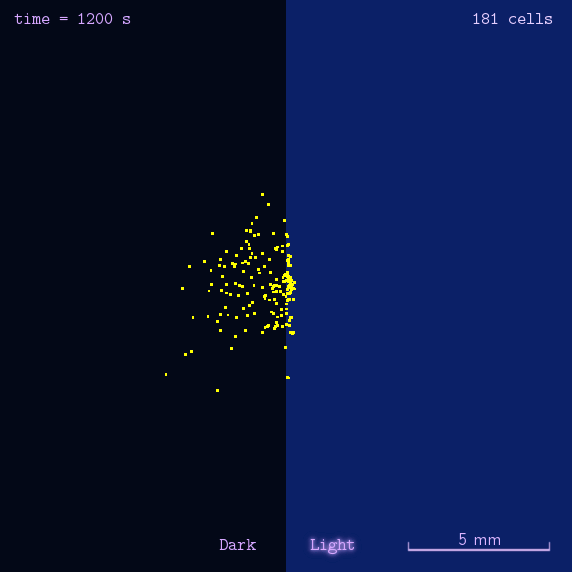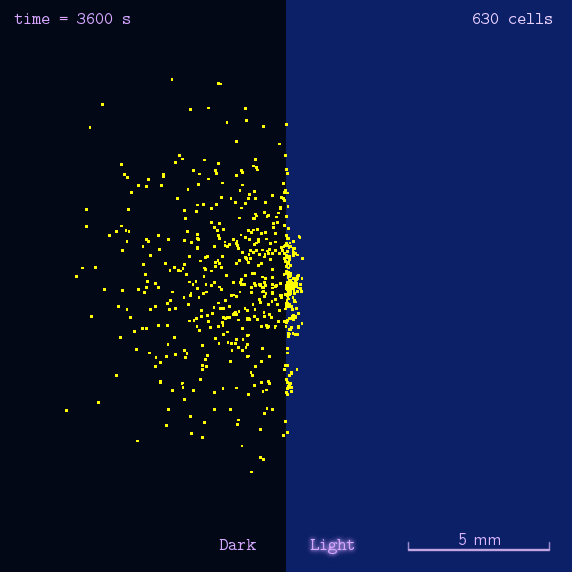
Phototaxis is a light-control bio-system, whose input is the space-distribution of light. By comparison with chemotaxis system, phototaxis has much advantages for application. (See Project Phototaxis) We have constructed a simple phototaxis system coupling our Luminesensor with the expression level of cheZ protein. In order to confirm the macro light-control to our phototaxis system, we used the Mean-field PDE model. Later we managed to confirm these phenomena by tracing each cells in Stochastic Simulation. We then managed to do the related experiments to prove the effect of light in this simple system.
On the way to the simulation of the macro population changing process, we developed a hexagonal-coordinate simulation environment for dynamic system on a continuous plane. Since the number of neighbors of a chunk unit in the hexagonal mesh is larger than that of the traditional quadratic mesh, the hexagonal mesh is much closer to an isotropic grid, which would reduce the error caused by anisotropic structure in traditional quadratic mesh. This simulation environment would be a useful prototype for future simulation of 2D dynamic systems, such as cellular movement on plate and pattern formation based on cell-cell communication.
Reference
- 1. Zoltowski, B.D., Crane, B.R.(2008). Light Activation of the LOV Protein Vivid Generates a Rapidly Exchanging Dimer. Biochemistry, 47: 7012: 7019
 "
"














