Team:NTNU Trondheim/Experiments and Results
From 2012.igem.org
(→Regulative LacI generator (BBa_K822004)) |
|||
| Line 11: | Line 11: | ||
</html> | </html> | ||
__TOC__ | __TOC__ | ||
| - | + | ==BioBrick parts== | |
| - | == | + | |
| - | + | ||
To make a genetic circuit releasing colicin as a response to a low oxygen level and a high lactate level, we needed several biobricks. For a detailed list of all biobricks present in our construct, see the [https://2012.igem.org/Team:NTNU_Trondheim/Parts parts page]. | To make a genetic circuit releasing colicin as a response to a low oxygen level and a high lactate level, we needed several biobricks. For a detailed list of all biobricks present in our construct, see the [https://2012.igem.org/Team:NTNU_Trondheim/Parts parts page]. | ||
| Line 28: | Line 26: | ||
This page will focus on the biobricks we have made, how we made them, and how we have characterized them to show that they work. | This page will focus on the biobricks we have made, how we made them, and how we have characterized them to show that they work. | ||
| - | + | ==Colicin (<partinfo>BBa_K822002</partinfo>)== | |
| - | + | ||
Colicin is the protein we have chosen as toxin in our bacterial anti-cancer-kamikaze device. We amplified the brick using <partinfo>BBa_K150009</partinfo> as template. The brick contains protein coding sequences both for Colicin E1 and for colicin immunity protein. The following primers were used: | Colicin is the protein we have chosen as toxin in our bacterial anti-cancer-kamikaze device. We amplified the brick using <partinfo>BBa_K150009</partinfo> as template. The brick contains protein coding sequences both for Colicin E1 and for colicin immunity protein. The following primers were used: | ||
| Line 64: | Line 61: | ||
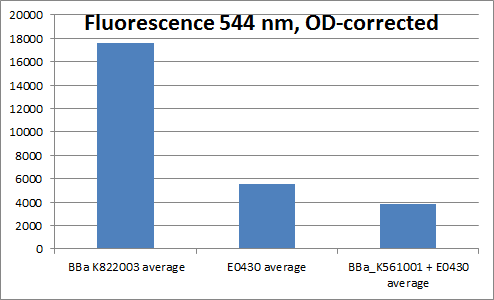
| - | + | ==YFP generator (<partinfo>BBa_K822003</partinfo>)== | |
We made the YFP generator to verify that the YFP coding sequence worked, in order to use it in experiments verifying that the Vgb promoter worked. The YFP generator was cloned together from a constitutive promoter + RBS (<partinfo>BBa_K081005</partinfo>), YFP (<partinfo>BBa_E0030</partinfo>) and a double terminator (<partinfo>BBa_B0015</partinfo>). | We made the YFP generator to verify that the YFP coding sequence worked, in order to use it in experiments verifying that the Vgb promoter worked. The YFP generator was cloned together from a constitutive promoter + RBS (<partinfo>BBa_K081005</partinfo>), YFP (<partinfo>BBa_E0030</partinfo>) and a double terminator (<partinfo>BBa_B0015</partinfo>). | ||
| Line 74: | Line 71: | ||
Even though we were not able to prove that the Vgb promoter works, we proved that the YFP generator works, as the fluorescence divided by OD is much higher for cells containing this biobrick, than for example for cells containing plasmids with only YFP. | Even though we were not able to prove that the Vgb promoter works, we proved that the YFP generator works, as the fluorescence divided by OD is much higher for cells containing this biobrick, than for example for cells containing plasmids with only YFP. | ||
| - | + | ==Regulative LacI generator (<partinfo>BBa_K822004</partinfo>)== | |
We made this brick in an effort to improve an already existing biobrick. The brick we wanted to improve was <partinfo>BBa_K292006</partinfo>. The NTNU iGEM team 2011 tried to use this brick in their stress sensor, but did not get it to work. They also tried to test-cut it and investigate the fragments using gel electrophoresis, however the resulting fragments were not as expected. This is why we thought of this biobrick as a suitable candidate for improvement. | We made this brick in an effort to improve an already existing biobrick. The brick we wanted to improve was <partinfo>BBa_K292006</partinfo>. The NTNU iGEM team 2011 tried to use this brick in their stress sensor, but did not get it to work. They also tried to test-cut it and investigate the fragments using gel electrophoresis, however the resulting fragments were not as expected. This is why we thought of this biobrick as a suitable candidate for improvement. | ||
| Line 88: | Line 85: | ||
|} | |} | ||
| - | + | ==lld promoter + RBS from ''E.coli'' (<partinfo>BBa_K822000</partinfo>)== | |
The two criteria we wanted fulfilled to initiate lysis and subsequent release of colicin were a low oxygen level and a high lactate level. A promoter activated by low oxygen level was already present in the registry (microaerobic Vgb promoter, <partinfo>BBa_K561001</partinfo>), but we found no suitable lactate-induced promoter. Therefore, we decided to convert the promotor regulating the lldPRD operon in ''E.coli'' into a biobrick, and to use this biobrick in our project [[http://www.ncbi.nlm.nih.gov/pubmed/18263722 1]]. | The two criteria we wanted fulfilled to initiate lysis and subsequent release of colicin were a low oxygen level and a high lactate level. A promoter activated by low oxygen level was already present in the registry (microaerobic Vgb promoter, <partinfo>BBa_K561001</partinfo>), but we found no suitable lactate-induced promoter. Therefore, we decided to convert the promotor regulating the lldPRD operon in ''E.coli'' into a biobrick, and to use this biobrick in our project [[http://www.ncbi.nlm.nih.gov/pubmed/18263722 1]]. | ||
| Line 111: | Line 108: | ||
| - | + | ==ldhA promoter + RBS from ''C.glutamicum'' (<partinfo>BBa_K822001</partinfo>)== | |
We also amplified the ldhA promoter from ''Corynebacterium glutamicum''. This has similar properties as the lld promoter from ''E.coli'', so this promoter was also a candidate to being used as the lactate inducable promoter in our project. | We also amplified the ldhA promoter from ''Corynebacterium glutamicum''. This has similar properties as the lld promoter from ''E.coli'', so this promoter was also a candidate to being used as the lactate inducable promoter in our project. | ||
Revision as of 23:10, 26 September 2012
 "
"